The CircHAS2/RPL23/MMP9 Axis Facilitates Brain Tumor Metastasis
1Department of Neurosurgery, First Hospital of Xinjiang Medical University, Urumqi, Xinjiang 830054, China
2Department of Anesthesiology, Sun Yat-sen Memorial Hospital of Sun Yat-sen University, Guangzhou, Guangdong 510120, China
3Medical Research Center of Shenshan Medical Center, Memorial Hospital of Sun Yat-Sen University, Shanwei, Guangdong 516600, China
4Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Guangdong-Hong Kong Joint Laboratory for RNA Medicine, Medical Research Center, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, China
5Nanhai Translational Innovation Center of Precision Immunology, Sun Yat-Sen Memorial Hospital, Foshan 528200, China
aThese authors contributed equally to this work.
*Correspondence to: Professor Phei Er Saw, PhD, 107 Yan Jiang West Road, Guangzhou, Guangdong, China, Tel: +86 13926401219. E-mail: caipeie@mail.sysu.edu.cn; Yongxin Wang, MD, 137 Li Yu Shan Road, Urumqi, Xinjiang, China, Tel: +86-15899225560. E-mail: xjdwyx2000@sohu.com
Received: 6 August 2023; Revised: 12 September 2023; Accepted: 20 November 2023; Published Online: 2 January 2024
Cite this paper:
Qiang Fu, Haojie Yang, Jingxuan Huang, Fan Liu, Yanni Fu, Phei Er Saw, and Yongxin Wang. The CircHAS2/RPL23/MMP9 Axis Facilitates Brain Tumor Metastasis. BIO Integration 2024; 5: e999.
DOI: 10.15212/bioi-2023-0013. Available at: https://bio-integration.org/
Download citation
© 2024 The Authors. This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See https://bio-integration.org/copyright-and-permissions/
Abstract
Background: Circular RNAs (circRNAs) regulate tumor development by interacting with microRNAs. However, limited research has been conducted on the roles of circRNAs in gliomas. Therefore, we sought to demonstrate the function and molecular mechanism of circHAS2 in gliomas.
Methods: CircHAS2, hsa-miR-508-3p, RPL23, and MMP9 mRNA levels were assessed with qRT-PCR. RPL23 and MMP9 protein levels were determined with western blotting and immunohistochemical staining. Glioma cell migration and invasion were assessed with Transwell assays. The interaction between hsa-miR-508-3p and circHAS2 or RPL23 was predicted with RNAhybrid and miRanda, and confirmed through luciferase reporter assays. The effects of circHAS2 on glioma cells were demonstrated in a nude mouse orthotopic xenograft glioma model.
Results: We computationally analyzed the differentially expressed circRNAs in glioma tissues by using the GEO database. The screening indicated that circHAS2 was located primarily in the cytoplasm. Functionally, silencing of circHAS2 inhibited glioma migration and invasion. Mechanically, hsa-miR-508-3p was identified as a downstream target of circHAS2. CircHAS2 was found to regulate RPL23 and influence MMP9 via hsa-miR-508-3p, thereby promoting glioma migration and invasion. Moreover, inhibition of circHAS2 impeded the progression of U87 glioma cells in vivo.
Conclusion: CircHAS2 regulates RPL23 and subsequent MMP9 expression by sponging hsa-miR508-3p in glioma cells.
Keywords
circHAS2, glioma, hsa-miR-508-3p, MMP9, RPL23.
Introduction
Glioma, a difficult-to-cure primary malignant brain tumor, has the highest incidence among central nervous system tumors [1]. Despite advanced treatments, including surgery, chemotherapy, and radiotherapy, the prognosis for glioma remains dismal [2], and targeted therapies are lacking. Because of its complicated pathogenesis, diagnostic or therapeutic decisions are highly difficult to make, and remain debated [3]. Therefore, identifying the pivotal target in glioma and elucidating its role would contribute to enhancing patient outcomes. Circular RNAs (circRNAs)—covalent, closed-loop forms of non-coding RNA—are characterized by the absence of a 3´-end polyadenylated (poly[A]) tail or 5´-end cap structure; consequently, these RNAs are less susceptible to degradation by exonucleases and more stable than their linear RNA counterparts [4]. The circRNAs discovered to date are derived primarily from gene exons and are abundant in the cytoplasm in eukaryotic cells [5]. Interestingly, circRNAs contain many miRNA-binding sites that enable them to act as miRNA sponges, and consequently inhibit the regulatory effects of miRNAs and enhance target-gene expression [6]. For example, Li et al. have verified that hsa_circ_0081534 facilitates nasopharyngeal carcinoma cell proliferation and metastasis through regulating the miR-508-3p/FNX axis [7]. Many studies have confirmed that various circRNAs are involved in the regulation of tumor formation and development by interacting with miRNAs [4, 8, 9]. However, because studies regarding the roles of circRNAs in glioma remain limited, continued exploration is warranted.
In this study, through database and qRT-PCR analysis, we identified circHAS2, a circRNA originating from human hyaluronan synthase 2 (HAS2). Its ID in circBase is hsa_circ_0005015. This circRNA is highly expressed in glioma. In vitro, silencing of circHAS2 inhibited glioma migration and invasion. Through luciferase reporter gene assays, we demonstrated that hsa-miR-508-3p binds circHAS2. Mechanistically, circHAS2 was found to target and regulate RPL23 expression via sponging hsa-miR-508-3p; moreover, RPL23 may upregulate the expression of MMP9. In vivo, depletion of circHAS2 inhibited U87 glioma cell proliferation, and the expression of RPL23 and MMP9. This study provides the first evidence that circHAS2 may promote glioma migration and invasion, thus enriching the study of circRNA function in glioma and providing a potential target for glioma treatment.
Results
circHAS2 is upregulated in brain tumor tissue
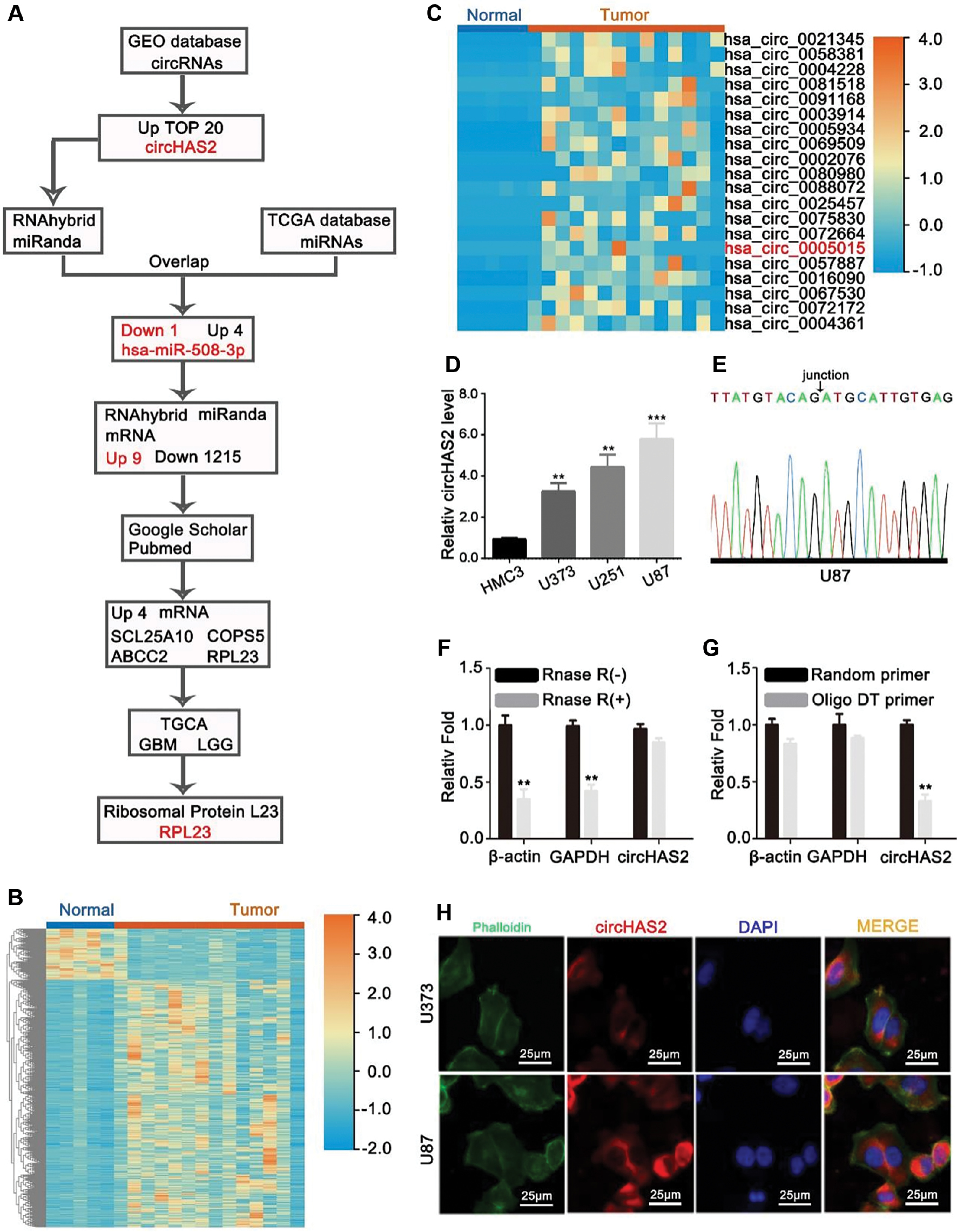
The flowchart in Figure 1A provides an overview of the analysis process. First, we analyzed brain tumor circRNA sequencing data from the GEO database. We selected the top 20 upregulated circRNAs for validation. Among them, circHAS2 was demonstrated to be significantly increased. We used databases to predict miRNAs that could potentially bind circHAS2 and simultaneously analyzed brain tumor miRNA sequencing data from TCGA database. Through intersecting the results, we identified one downregulated miRNA and four upregulated miRNAs. Given that the literature has reported a negative correlation between circRNA and miRNA expression levels [10], we selected hsa-miR-508-3p, which was downregulated in brain tumors, for further investigation. Next, through database predictions, we identified nine upregulated mRNAs and 1,215 downregulated mRNAs that might potentially bind has-miR-508-3p. Because the literature has indicated that miRNA expression is inversely associated with its target genes [11], we selected the nine upregulated mRNAs for further research. Using these nine gene names and “tumor” as keywords, we conducted a literature search of the Google Scholar and PubMed databases, and determined that only four of these genes had been reported to function in tumors. Therefore, we further investigated these four genes. TCGA database results demonstrated significant RPL23 upregulation in brain tumors and multiple other tumors. Consequently, we selected RPL23 as a candidate gene for subsequent validation. We first used the GEO database (Figure 1B) to identify circRNAs differentially expressed in glioma. The top 20 circRNAs were screened, and a heatmap was generated (Figure 1C). Subsequently, qRT-PCR confirmed the higher abundance of circHAS2 in brain tumor cell lines than controls, and identified circHAS2 as a potential gene for further study. We observed upregulation of circHAS2 in brain tumor cell lines compared with normal brain glial cells (Figure 1D). Sanger sequencing confirmed the base sequence at circHAS2-binding sites (Figure 1E). Because of their loop-like structure lacking a poly(A) tail, circRNAs including circHAS2 are resistant to RNase digestion and are more stable than linear RNA molecules in vitro [12] (Figure 1F). For all circRNAs transcribable by random primers, we designed both random and oligo DT primers for circHAS2 synthesis. As expected, only the random primers resulted in successful transcription of circHAS2, thus confirming its loop structure (Figure 1G). Finally, fluorescence in situ hybridization (FISH) experiments revealed that circHAS2 was located primarily in the cytoplasm in glioma cells (Figure 1H, Supplementary Figure 1).
Figure 1 circHAS2 is a differentially expressed circRNA in brain tumors, as identified through a database search. (A) Flowchart of the study. (B) Heatmap of differentially expressed circRNAs in brain tumors and normal brain tissue. (C) Heatmap of the top 20 circRNAs differentially expressed in brain tumors versus normal brain tissue. (D) Expression of circHAS2 in brain tumor cell lines. (E) The base sequences at the junction of circHAS2, identified by Sanger sequencing. (F) Changes in the expression levels of β-actin, GAPDH, and circHAS2 after RNase treatment, as detected by qRT-PCR. (G) Changes in expression levels detected by qRT-PCR after synthesis of β-actin, GAPDH, and circHAS2 cDNA with random primers and oligo DT primers. (H) Distribution of circHAS2 expression in brain tumor cells, identified by FISH. * p < 0.05, ** p < 0.01, *** p < 0.001.
Silencing of circHAS2 inhibits migration and invasion of U87 and U251 cells
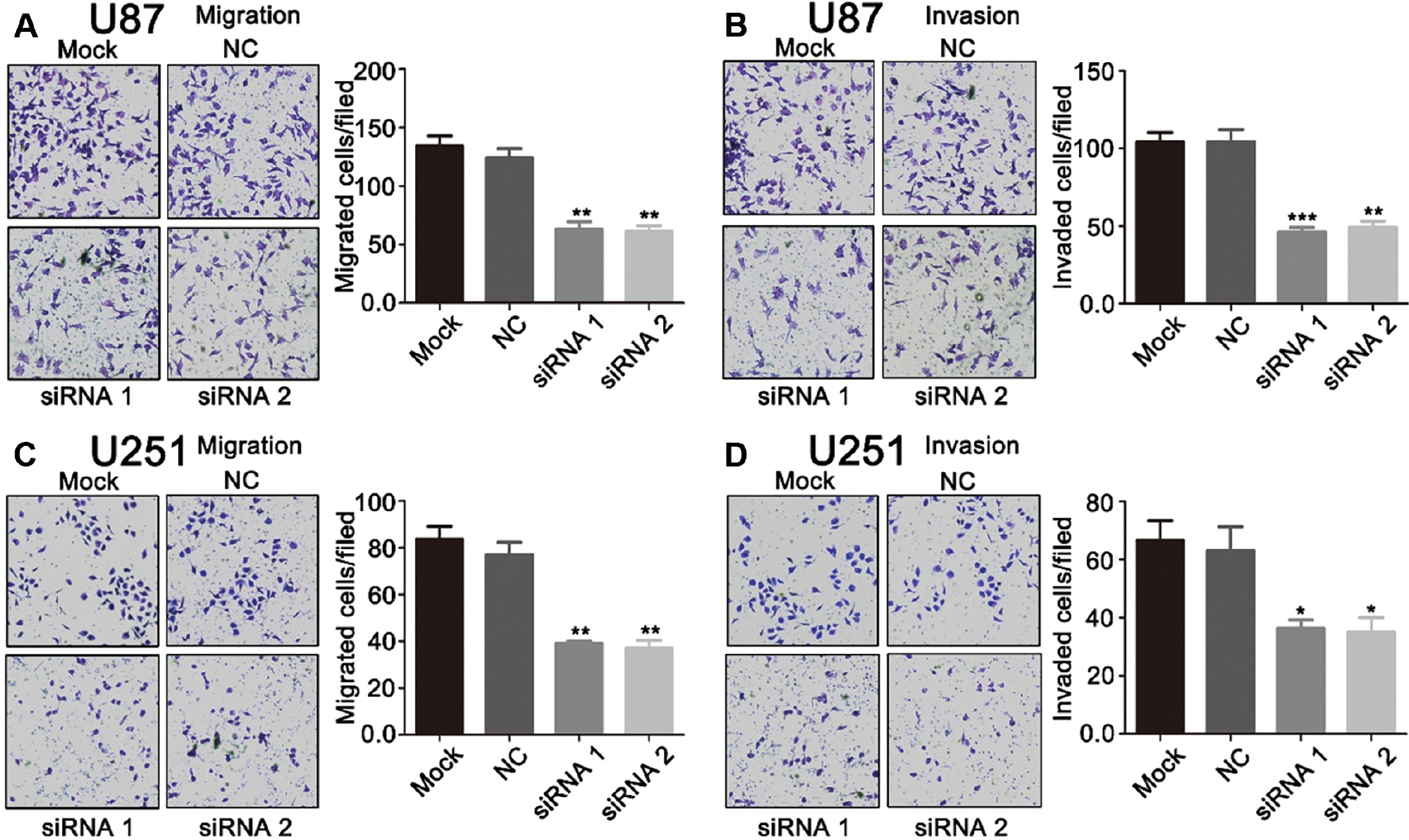
The expression of circHAS2 was higher in brain tumor cells than in normal human glial cells; therefore, we hypothesized that this circRNA might act as an oncogene in brain tumors. To test this hypothesis, we designed siRNAs for circHAS2. Silencing of circHAS2 in the U87 cell line effectively silenced circHAS2 without interfering with the expression of the parental gene HAS2 (Supplementary Figure 2). The influence of circHAS2 on the migratory and invasive capacity of brain tumor cell lines was verified with Transwell assays. Silencing of circHAS2 significantly suppressed the migration and invasion of the brain tumor cell lines U87 (Figure 2A and B) and U251 (Figure 2C and D).
Figure 2 CircHAS2 functions in brain tumor migration and invasion. (A) U87 migration with or without circHAS2 siRNA transfection, detected with Transwell assays. (B) U87 invasion with or without circHAS2 siRNA transfection, detected with Transwell assays. (C) U251 migration with or without circHAS2 siRNA transfection, detected with Transwell assays. (D) U251 invasion with or without circHAS2 siRNA transfection, detected with Transwell assays. * p < 0.05, ** p < 0.01, *** p < 0.001.
circHAS2 exerts pro-tumor effects via sponging hsa-miR-508-3p
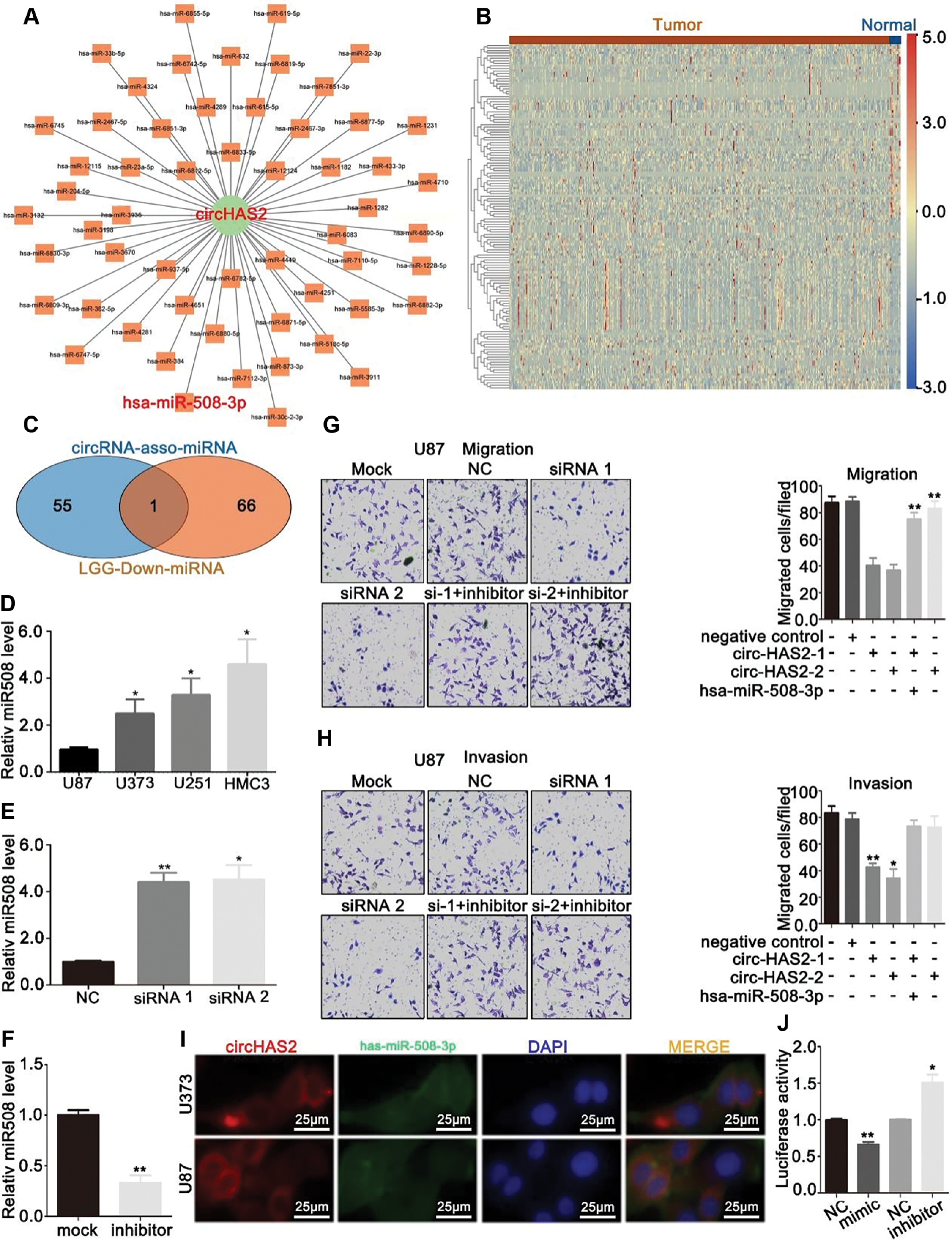
The localization of circRNAs dictates the mechanisms through which tumor biological processes are influenced [13]. CircRNAs in the nucleus predominantly bind transcription factors, whereas those in the cytoplasm associate with both RNA-binding proteins and miRNAs [14, 15]. Potential miRNAs that might bind circHAS2 were predicted with the RNAhybrid and miRNA databases (Figure 3A). Concurrently, the miRNAs differentially expressed between brain tumor tissues and normal brain tissues in the GEO database were analyzed, and a heatmap was generated (Figure 3B). The intersection of these two datasets was established, on the basis of the reported negative correlation between the expression of circRNAs and miRNAs [8, 16–20]. Subsequently, downregulated miRNAs in brain tumor tissues were selected and intersected with the potential miRNAs predicted to be bound by circHAS2 in the database, and one miRNA, hsa-miR-508-3p, was identified (Figure 3C).
Figure 3 CircHAS2 may act as a sponge for hsa-miR-508-3p. (A) Network map of miRNAs predicted to be sponged by circHAS2, on the basis of a database search. (B) Heatmap of differentially expressed miRNAs in brain tumors and normal brain tissues. (C) Venn diagram of downregulated miRNAs expressed in brain tumors and database-predicted miRNAs that bind circHAS2. (D) Expression of hsa-miR-508-3p in brain tumor cell lines. (E) Changes in the expression of hsa-miR-508-3p in U87 cells after silencing of circHAS2, as detected by qRT-PCR. (F) Changes in the expression of hsa-miR-508-3p in U87 cells after hsa-miR-508-3p inhibitor treatment, as detected by qRT-PCR. (G) Changes in the migratory capacity of U87 cells after transfection with circHAS2 siRNA and/or hsa-miR-508-3p inhibitor, on the basis of Transwell assays. (H) Changes in the invasion capacity of U87 cells after transfection with circHAS2 siRNA and/or hsa-miR-508-3p inhibitor, on the basis of Transwell assays. (I) Subcellular localization of circHAS2 and hsa-miR-508-3p in brain tumor cells, identified by FISH. (J). Possible sponging of hsa-miR-508-3p by circHAS2 in HEK293T cells, verified by luciferase reporter gene assays. NC, negative control of mimic and inhibitor, respectively. All data are represented as mean ± SD; * p < 0.05, ** p < 0.01.
We next performed qRT-PCR to quantify the expression of hsa-miR-508-3p, which was found to be lower in normal human microglia than brain tumor cells (Figure 3D). Interestingly, silencing of circHAS2 appeared to promote hsa-miR-508-3p expression (Figure 3E). To confirm the effects of circHAS2 and hsa-miR-508-3p on the migration and invasion of U87 brain tumor cells, we used a combination of hsa-circ 0005015 silencing and hsa-miR-508-3p inhibition (Figure 3F). Silencing hsa_circ_0005015 and promoting hsa-miR-508-3p expression partially reversed the migration (Figure 3G) and invasion (Figure 3H) of U87 cells. These findings indicated that hsa_circ_0005015 enhances the migration and invasion of brain tumors, potentially via hsa-miR-508-3p.
Additionally, fluorescence co-localization assays demonstrated that hsa_circ_0005015 and hsa-miR-508-3p co-localized in the cytoplasm (Figure 3I). We then co-transfected hsa-miR-508-3p mimics and inhibitors, along with luciferase reporter genes, into HEK293T cells. The transfection efficiency of the hsa-miR-508-3p mimic was validated by qRT-PCR (Supplementary Figure 3). Transfection of hsa-miR-508-3p mimics decreased the fluorescence intensity, whereas transfection of hsa-miR-508-3p inhibitors increased the fluorescence intensity, thereby further confirming that circHAS2 sponges hsa-miR-508-3p (Figure 3J).
Hsa-miR-508-3p upregulates RPL23 expression
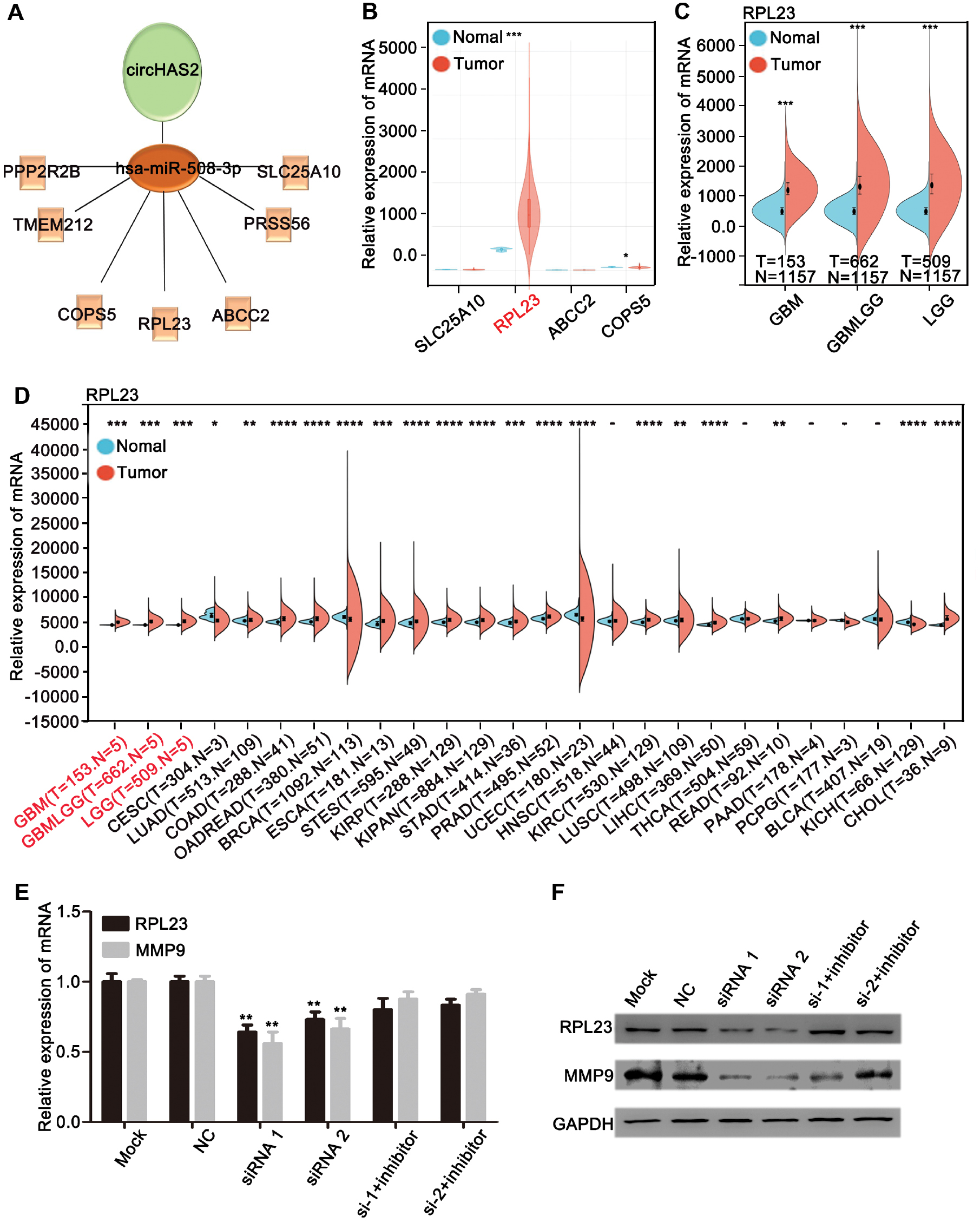
Previous research has indicated that miRNAs bind the 3’UTRs of target mRNAs and subsequently inhibit protein translation; moreover, miRNA expression is typically inversely correlated with its target-gene expression [21–26]. We therefore used the RNAhybrid and miRanda databases to predict mRNAs that might bind hsa-miR-508-3p. Seven mRNAs (PPPP2R2B, TMEM212, COPS5, RPL23, ABCC2, PRSS56, and SLC25A10) across the aforementioned databases were identified to potentially bind hsa-miR-508-3p (Figure 4A). As shown in Figure 2, we confirmed that circHAS2 functions as an oncogene in brain tumor cell lines. We next conducted a literature search to investigate the roles of these seven genes in tumors. Only four of these genes (SLC25A10, RPL23, ABCC2, and COPS5) had been documented (Figure 4B). Among those genes, only RPL23 had significantly higher expression in brain tumors than normal brain tissues, according to TCGA database, thus indicating that RPL23 was a potential a target gene for hsa-miR-508-3p (Figure 4C).
Figure 4 RPL23 may be a target gene of hsa-miR-508-3p. (A) Network of circHAS2, hsa-miR-508-3p, and its downstream target genes. (B) Relative expression levels of the SCL25A10, RPL23, ABCC2, and COPS5 genes, according to TCGA database. (C) Relative expression of the RPL23 gene in GBM, GBMLGG, and LGG, according to TCGA database. (D) Relative expression of the RPL23 gene in various tumor tissues, according to TCGA database. (E) Changes in the relative expression of the RPL23 gene in U87 cells transfected with circHAS2 siRNA and/or hsa-miR-508-3p inhibitor, as detected by qRT-PCR. (F) Changes in the relative expression of the RPL23 gene after transfection of U87 cells with circHAS2 siRNA and/or hsa-miR-508-3p inhibitor, as detected by western blotting. All data are represented as mean ± SD; * p < 0.05, ** p < 0.01, *** p < 0.01, **** p < 0.01.
Further analysis of TCGA database data revealed that RPL23 expression was significantly elevated in 21 of 26 types of tumors, including brain tumors, compared with normal tissues (Figure 4D), thereby suggesting a potential role of RPL23 in brain tumor development. Prior studies have reported that RPL23 promotes HCC metastasis by regulating MMP9 expression in HCC [27]. To clarify the regulatory effects of circHAS2 and hsa-miR-508-3p on RPL23 and MMP9, we conducted qRT-PCR (Figure 4E) and western blotting (Figure 4F). When circHAS2 was silenced along with the inhibition of hsa-miR-508-3p expression, a partial reversal of the decrease in RPL23 and MMP9 in U87 cells was observed. These results suggested that circHAS2 may regulate RPL23, thereby influencing MMP9 through hsa-miR-508-3p, and promoting the migration and invasion of brain tumors.
Silencing of circHAS2 inhibits metastasis of U87 brain tumor cells in vivo
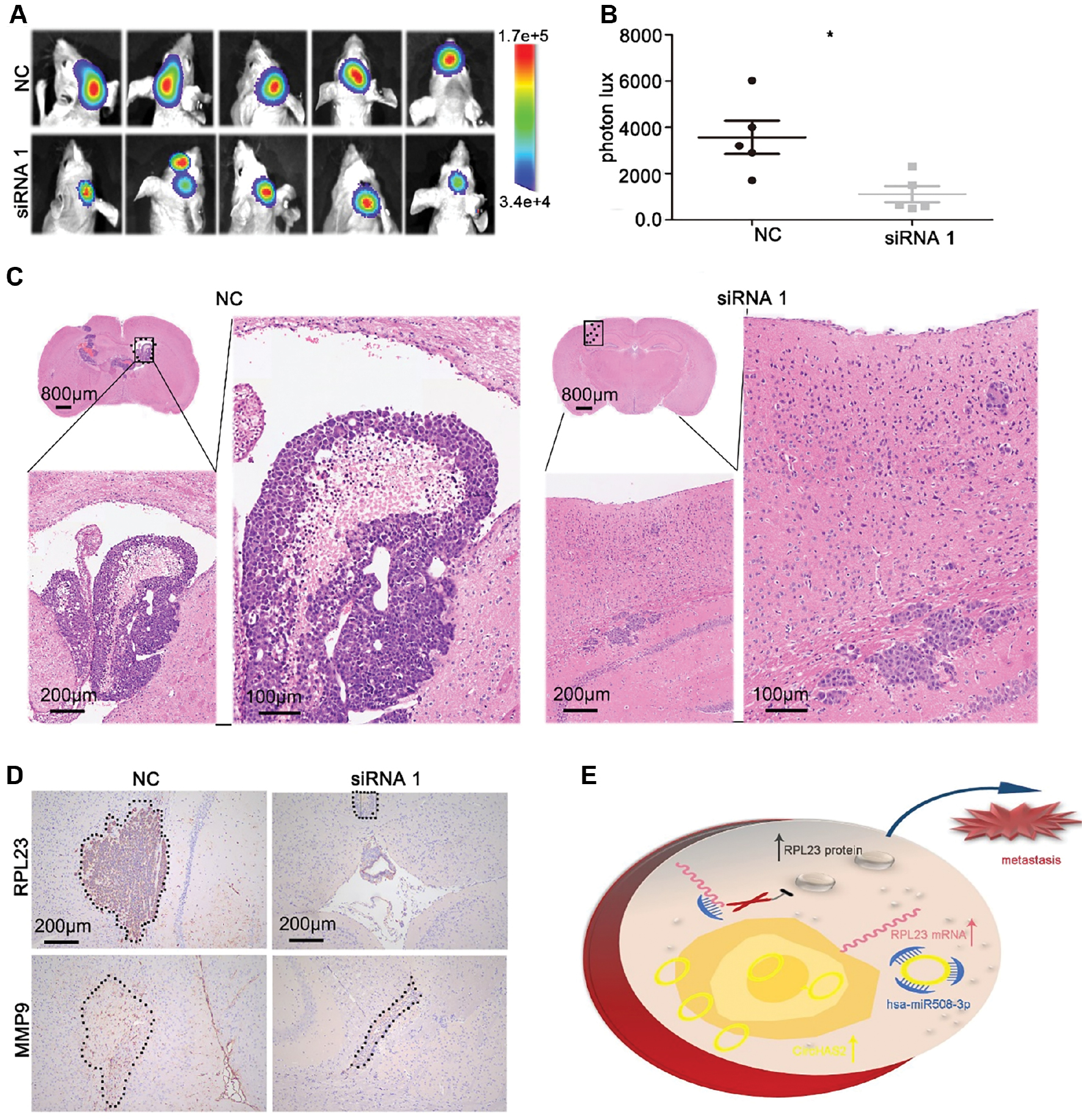
To further investigate the biological role of circHAS2 in brain tumor development, we transfected U87 cells with a lentivirus containing a luciferase reporter to generate U87-Luc cells. Subsequently, circHAS2 was silenced in U87-Luc cells with siRNA, and the modified cells were implanted into the brains of nude mice. Two weeks after implantation, an in vivo imaging spectrum system was used to track tumor foci formation in the mouse brains. Mice implanted with circHAS2-silenced cells exhibited lower fluorescence intensity in the tumor foci than the control mice. Representative immunohistochemistry staining of RPL23 and MMP9 in xenograft tumors from the circHAS2-silenced and control groups is shown in Figure 5D. The results demonstrated significant downregulation of the expression levels of RPL23 and MMP9 after circHAS2 silencing, in agreement with the findings from cellular experiments. In summary, our in vivo experiments indicated that circHAS2 inhibits the growth of U87 cells.
Figure 5 Silencing of circHAS2 restricts brain tumor metastasis in nude mice. (A) Effects of silencing of circHAS2 in U87 cells on the growth of xenograft tumors, observed with an in vivo imaging system. (B) Statistical graph of the fluorescence signal of the xenograft tumors and controls. (C) Representative images of HE staining of xenograft tumors in the circHAS2-silenced and control groups. (D) Representative images of immunohistochemistry staining of RPL23 and MMP9 in xenograft tumors in the circHAS2-silenced and control groups. (E) Schematic illustration of circHAS2’s role as an oncogene by sponging hsa-miR-508-3p, thereby promoting the expression of RPL23 and inducing glioma metastasis. All data are represented as the mean ± SD; * p < 0.05.
Discussion
In this study, we focused on the circRNA circHAS2, identified through bioinformatic analysis, because circRNAs have received substantial attention in cancer research, and the roles of circRNAs in cancer are critical [5]. Because circRNAs may be used as biomarkers for determining diagnosis and prognosis, tumorigenesis, proliferation, metastasis, angiogenesis, chemotherapy resistance, and radiotherapy resistance, we sought to explore the role of circRNA in glioma [28–31]. We found that circHAS2 was highly expressed in GBM cells and acts as an oncogene. In U87 cells, circHAS2 was found to regulate the expression of RPL23 and subsequently MMP9 by sponging hsa-miR-508-3p.
Previous studies have reported that circRNAs are involved in glioma progression. For example, circ_001350 promotes the malignant progression of glioma by adsorbing miRNA-1236 [32–40]. Furthermore, circ_0001801, circ_0067934, and circRNA FBXW7 have been demonstrated to be functional miRNA sponges in brain tumors [9, 34, 35]. Circ_0005015 (denoted circHAS2 herein) has been reported to promote the progression of diabetic retinopathy, and to act as an miR-519d-3p sponge, thereby upregulating levels of MMP2, XIAP, and STAT3 [36]. However, to our knowledge, the function and mechanism of circHAS2 in tumors, particularly in GBM, had not been reported. Interestingly, the expression of circHAS2 was found to be significantly up-regulated in glioma, and in vitro and in vivo experiments showed that circHAS2 may promote the metastasis of GBM cells. Many studies have indicated that circRNAs usually act as miRNA sponges, in an important molecular mechanism of regulating tumor progression [5]. Therefore, we sought to explore whether circHAS2 might also affect the biological function of glioma cells through such a mechanism.
In our study, hsa-miR-508-3p was predicted to be a promising target miRNA of circHAS2 via bioinformatics tools. Subsequently, the binding relationship was demonstrated with luciferase reporter gene assays [35–39]. For example, hsa-miR-508-3p is an important factor for the mesenchymal subtype and a key prognostic biomarker in ovarian cancer [39]. In addition to regulating the target gene by itself, the-miR-508-3p can also be sponged by circRNA, thus affecting tumor occurrence and development. Han et al. have reported that circHIPK3 modulates CXCL12 by spongithehsa-miR-508-3p; this mechanism may explain why circHIPK3 induces the proliferation and metastasis of ccRCC [38].
Interestingly, when we silenced the expression of circHAS2, the metastasis of GBM cells was inhibited, and the expression of the hsa-miR-508-3p target gene RPL23 decreased. Human ribosomal protein L23 (RPL23), and RNA Binding Protein 9 (RBP) are involved in the progression of various tumors, including HCC and lung cancer [40]. Yang and co-workers have demonstrated that RPL23 induces HCC metastasis by stabilizing MMP9 mRNA and upregulating MMP9 [27]. The matrix metalloproteinase family (MMP) mediates the degradation of the extracellular matrix, and MMP is associated with cancer cell metastasis [41]. MMP9, a member of the MMP family, plays an important role in BGM metastasis [42]. Importantly, RBP and HuR upregulate MMP9 levels by regulating MMP9 mRNA stability [27, 43].
In summary, we demonstrated that circHAS2 is an oncogene in GBM. No studies to date have elucidated the importance of circHAS2 as a key factor promoting glioma metastasis. On the basis of our results, circHAS2 regulates glioma migration and invasion by sponging hsa-miR-508-3p, and RPL23 and MMP9 are also involved in this regulatory mechanism. Thus, our findings may lead to a new therapeutic scheme.
Materials and methods
Data collection and application
Differential expression analysis of circRNAs and miRNAs was performed on datasets GSE159260 and GSE91014 from the GEO database. For circRNA, the fastq data of GSE159260 were downloaded. Quality control was conducted with fastQC. For all fastq files, the clean reads were aligned to the reference genome hg38 with bowtie2 (version 2.4.4). After alignment, find_circ (version 2) and samtools (version 1.13) were used to extract reads and convert the reads unmapped to the reference genome. All unmapped reads were used to predict candidate circRNAs by using find_circ (version 2). Bedtools (version 2.30) was used to annotate the circRNAs to circBase and identify their related host genes. DESeq2 was used to perform differential expression analysis of circRNAs.
For miRNAs, the raw data from GSE91014 were downloaded. The R package Affy (version 1.64.0) was used to preprocess and normalize the raw data. the R package limma (version 3.42.2) was used to perform differential analysis of miRNAs. miRNAs that reached the cut-off threshold value of p value < 0.05 and an absolute value of the fold change >1.5 were considered differentially expressed miRNAs.
Differentially expressed circRNAs and differentially expressed miRNAs were detected from the GEO database. CircRNA IDs and sequences were collected from circBase. The miRNA sequences were collected from miRbase. RNAhybrid (version 2.2) and miRanda (version 3.3a) were used to predict the possible miRNAs binding the identified circRNA, and the possible mRNA targets of the identified miRNAs.
Cytoscape (version 3.9.1) was used to build the circRNA-miRNA-mRNA network. Research on nine potential target genes of miRNAs in tumors were explored in Google Scholar and PubMed, with keywords including SCCL25A10 cancer, COPS5 cancer, ABCC2 cancer, and RPL23 cancer. Subsequently, we identified genes studied extensively in tumors during the past 5 years. SCCL25A10, COPS5, ABCC2, and RPL23 were identified as possible candidates for further exploration. TCGA database (https://portal.gdc.cancer.gov/) was used to analyze the relative expression of the four selected genes in brain tumor and normal brain tissues, and the levels of RPL23 in other cancers were analyzed on the basis of TCGA database (https://portal.gdc.cancer.gov/).
Cell lines culture and transfection
The HEK293T, HMC3, U251, and U87 cell lines were purchased from the American Type Culture Collection, USA). All cell lines were cultured in a 37°C humidified 5% CO2 incubator with the relevant media. U251 and U87 were transfected with siRNA (Genepharma, Shanghai, China) with iMAX (Invitrogen, MA, USA). The sequences of siRNA 1 and siRNA 2 of circHAS2 are provided in Supplementary Table 1. The hsa-miR-508-3p inhibitor, mimics, and negative control (NC), purchased from (Ribobio, Guangzhou, China), were transfected into U251 and U87 cells with a Ribo FECT CP transfection Kit (Ribobio), according to the transfection procedures described on the company’s official website (https://www.ribobio.com/).
Luciferase reporter assays
A dual luciferase reporter system kit was purchased from Beyotime (Shanghai, China). HEK293T cells were seeded in 96-well plates and transfected with PRL-TK-pMIR-circHAS2 or PRL-TK-pMIR-508 3´UTR, and hsa-miR-508-3p mimics or miR-NC at 37°C for 48 h. The relative firefly and Renilla luciferase activity was measured according to the kit protocol.
RNA fluorescence in situ hybridization
A circRNA FISH kit and miRNA FISH kit were purchased from Ribobio (China), and all procedures were performed according to the manufacturer’s instructions. Briefly, U251 and U87 cells were cultured to an appropriate density, fixed with 4% paraformaldehyde in the plates, washed with PBS, and incubated with pre-hybridization solution. The cells were subsequently incubated with FISH probe at 37°C for 8 h. The plates were washed with 4× SSC containing 0.1% Tween-20 three times, then with 2× SSC, 1× SSC, and PBS. Cell nuclei were stained with DAPI before fluorescence detection (IX83, OLYMPUS, Japan). The nucleotide sequence of circHAS2 and the probe sequence of circHAS2 are presented in Supplementary Table 2.
Immunofluorescence
U251 and U87 cells were cultured to an appropriate density, fixed with 4% paraformaldehyde in the plates, washed with PBS, and incubated with primary antibodies to G-actin (Cell Signaling Technology, Boston, USA) at 4°C for 8 h. The cells were incubated with FITC conjugated secondary antibody (Invitrogen, Carlsbad, USA) at 37°C for 2 h, and subsequently incubated with DAPI. The images were detected with an IX83 microscope (OLYMPUS, Japan).
Western blotting
The collected proteins were resolved with 8% SDS-PAGE (Bio-Rad, Berkely, USA), and the separated proteins were transferred to a PVDF membrane (Merck Millipore, Germany), which was incubated with primary antibodies to human RPL23 (Thermo Fisher, Waltham, USA), MMP9 (Cell Signaling Technology), or GAPDH (Cell Signaling Technology) at 4°C for 8 h. The membrane was washed with 1× TBST containing 0.1% Tween-20 three times, then incubated with secondary antibodies (Cell Signaling Technology) at room temperature for 2 h, and examined with a chemiluminescence detection machine (Thermo Fisher Scientific).
Immunohistochemistry
Brain tissue was immersed in 4% paraformaldehyde, embedded in paraffin, and serially sectioned. The brain slides were incubated with primary antibodies to human RPL23 (Thermo Fisher) and MMP9 (Cell Signaling Technology) at 4°C for 8 h. The brain slides were washed with 1× PBS three times, then incubated with reagent from an HRP-labeled streptavidin kit (Dako, Denmark) at room temperature for 10 min. Subsequently, DAB was used to detect immunocomplexes, and the cell nuclei were stained with hematoxylin. Images were acquired with an IX83 microscope (OLYMPUS, Japan).
Transwell assays
A total of 1×105 cells were added to an upper Transwell chamber (Corning, New York, USA) with 200 μl serum free medium, and 500 μl medium with 10% FBS was added to the lower Transwell chamber. Cells were cultured 24 h (migration) or 48 h (invasion). Cell invasive mobility was tested on the upper chamber precoated with Matrigel (Thermo Fisher). The upper chamber was washed with PBS, a cotton swab was used to remove cells that had not migrated, and the cells were fixed in 4% paraformaldehyde. All cells were stained with crystal violet and counted.
RNase R assays and nuclear and cytoplasmic RNA collection
Total RNA was treated with 4 U RNase R/μg RNA (Geneseed, Guangzhou, China) or 0.2 μl RNase free water/ug (Beyotime, China) at 37°C for 30 min. Subsequently, the treated RNA was used to examine the levels of circHAS2, β-actin, and GAPDH via qRT-PCR. The nuclear and cytoplasmic RNA fractions were extracted with a PARAIS kit (Invitrogen, USA). To evaluate the relative expression levels of circHAS2 in the cytoplasm and nucleus, β-actin was employed as the internal reference for cytoplasmic RNA, while U6 served as the internal reference for nuclear RNA. The circHAS2, β-actin, GAPDH, and U6 primer sequences are presented in Supplementary Table 3.
Real-time quantitative PCR
The qRT-PCR was performed with Bio-Rad CFX Connect system (USA). Total RNA was collected with Trizol (Beyotime). The RNA was translated into cDNA with an miRNA Reagent kit (Riobio) or CircRNA Reagent kit (Takara, Dalian, China). The relative level of miRNA was measured with Riobio SYBR Green PCR kits (Riobio), and the expression of circRNA was examined with a SYBR Premix Ex TaqIIkit (Takara). Each detection was conducted with three replicate wells. All procedures were performed according to the manufacturer’s instructions. U6 served as the miRNA internal control, and β-actin served as the circRNA and mRNA internal control. All data were analyzed with 2−ΔΔCT.
Xenograft mouse models
The animal experiment was authorized by the Sun Yat-sen University Laboratory Animal Care and Use Committee. BALB/C nude mice 4 weeks old were purchased from Sun Yat-Sen University Laboratory Animal Center and used to establish intracranial brain tumor models as previously reported. The pLv-Luc lentivirus was transduced into the U87MG cell line to generate a bioluminescent U87MG luciferase puromycin (U87MG-Luc) expressing cell line. U87MG-Luc cells were incubated with siRNA (Genepharma, China) against circHAS2 with iMAX (Invitrogen, USA). Two transfections with siRNA were performed consecutively. A total of 1×106 cells/μL in 5 μL PBS were injected into the mouse brain via the skull, and each mouse was imaged 2 weeks later with an IVIS Spectrum series instrument (Perkin Elmer, USA) after intraperitoneal injection of 90 μL of d-luciferin (15 mg/ml, Yeasen, Shanghai, China) 9 minutes before photography. Brains were collected and fixed in paraformaldehyde for subsequent experiments after imaging.
Statistical analysis
Continuous variables are reported as means ± standard error, and categorical variables are reported as numbers. The Kruskal-Wallis test and Mann-Whitney u test were used to evaluate significant differences between groups. All statistical tests were performed in GraphPad Prism version 6.5 (GraphPad Software Inc, San Diego, USA). P < 0.05 was considered statistically significant.
Acknowledgement
The authors would like to express their sincere appreciation to Yuanqi Feng for his support in bioinformation technology.
Declaration of competing interests
The authors declare no conflicts of interest.
Funding
This study was supported by the National Natural Science Foundation of Xinjiang, Uygur Autonomous Region, China (grant No. 2021D01C312) and Guangzhou Province Basic Research Fund (202201020576).
Ethical approval
This study was performed in line with the principles of the Sun Yat-Sen University. Approval was granted by the Institutional Animal Care and Use Committee of Sun Yat-Sen University (SYSU-IACUC-2019-B748).
Availability of data and materials
Please contact the corresponding author for data requests.
Consent for publication
Not applicable.
Abbreviations
| GBM | Glioblastoma Multiforme |
| GBMLGG | Glioblastoma and Lower Grade Glioma |
| LGG | Lower Grade Glioma |
| CESC | Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma |
| LUAD | Lung Adenocarcinoma |
| COAD | Colon Adenocarcinoma |
| OADREAD | Ovarian Adenocarcinoma and Endometrioid |
| BRCA | Breast Cancer Susceptibility Gene |
| ESCA | Esophageal Carcinoma |
| STES | Stomach and Esophageal Carcinoma |
| KIRP | Kidney Renal Papillary Cell Carcinoma |
| KIPAN | Kidney Pan-Cancer |
| STAD | Stomach Adenocarcinoma |
| PRAD | Prostate Adenocarcinoma |
| UCEC | Uterine Corpus Endometrial Carcinoma |
| HNSC | Head and Neck Squamous Cell Carcinoma |
| KIRC | Kidney Renal Clear Cell Carcinoma |
| LUSC | Lung Squamous Cell Carcinoma |
| LIHC | Liver Hepatocellular Carcinoma |
| THCA | Thyroid Carcinoma |
| READ | Rectum Adenocarcinoma |
| PAAD | Pancreatic Adenocarcinoma |
| PCPG | Pheochromocytoma and Paraganglioma |
| BLCA | Bladder Urothelial Carcinoma |
| KICH | Kidney Chromophobe |
| CHOL | Cholangiocarcinoma |
Supplementary Figures and Tables
Click here to download Supplementary Files.
References
- An M, Zheng H, Huang J, Lin Y, Luo Y, et al. Aberrant nuclear export of circNCOR1 underlies SMAD7-mediated lymph node metastasis of bladder cancer. Cancer Res 2022;82:2239-53. [PMID: 35395674 DOI: 10.1158/0008-5472.CAN-21-4349]
- Anjum K, Shagufta BI, Abbas SQ, Patel S, Khan I, et al. Current status and future therapeutic perspectives of glioblastoma multiforme (GBM) therapy: a review. Biomed Pharmacother 2017;92:681-9. [PMID: 28582760 DOI: 10.1016/j.biopha.2017.05.125]
- Cao LQ, Yang XW, Chen YB, Zhang DW, Jiang XF, et al. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer 2019;18:148. [PMID: 31656200 DOI: 10.1186/s12943-019-1075-2]
- Chen B, Dragomir MP, Yang C, Li Q, Horst D, et al. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Ther 2022;7:121. [PMID: 35418578 DOI: 10.1038/s41392-022-00975-3]
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381-8. [PMID: 25746834 DOI: 10.1080/15476286.2015.1020271]
- Chen WL, Jiang L, Wang J-S, Liao CX. Circ-0001801 contributes to cell proliferation, migration, invasion and epithelial to mesenchymal transition (EMT) in glioblastoma by regulating miR-628-5p/HMGB3 axis. Eur Rev Med Pharmacol Sci 2019;23:10874-85. [PMID: 31858556 DOI: 10.26355/eurrev_201912_19791]
- Chi Y, Zheng W, Bao G, Wu L, He X, et al. Circular RNA circ_103820 suppresses lung cancer tumorigenesis by sponging miR-200b-3p to release LATS2 and SOCS6. Cell Death Dis 2021;12:185. [PMID: 33589592 DOI: 10.1038/s41419-021-03472-7]
- Claus EB, Walsh KM, Wiencke JK, Molinaro AM, Wiemels JL, et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus 2015;38:E6. [PMID: 25552286 DOI: 10.3171/2014.10.FOCUS12367]
- Cui YH, Feng QY, Liu Q, Li HY, Song XL, et al. Posttranscriptional regulation of MMP-9 by HuR contributes to IL-1β-induced pterygium fibroblast migration and invasion. J Cell Physiol 2020;235:5130-40. [PMID: 31691974 DOI: 10.1002/jcp.29387]
- Liu ZH, Zhou Y, Liang GH, Ling Y, Tan WG, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis 2019;10:55. [PMID: 30670688 DOI: 10.1038/s41419-018-1287-1]
- Xiong DD, Dang YW, Lin P, Wen DY, He RQ, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med 2018;16:220. [PMID: 30092792 DOI: 10.1186/s12967-018-1593-5]
- Du Y, Wei N, Ma R, Jiang S, Song D. A miR-210-3p regulon that controls the Warburg effect by modulating HIF-1α and p53 activity in triple-negative breast cancer. Cell Death Dis 2020;11:731. [PMID: 32908121 DOI: 10.1038/s41419-020-02952-6]
- Guo SJ, Zeng HX, Huang P, Wang S, Xie C-H, et al. MiR-508-3p inhibits cell invasion and epithelial-mesenchymal transition by targeting ZEB1 in triple-negative breast cancer. Eur Rev Med Pharmacol Sci 2018;22:6379-85. [PMID: 30338806 DOI: 10.26355/eurrev_201810_16050]
- Han B, Shaolong E, Luan L, Li N, Liu X. CircHIPK3 promotes clear cell renal cell carcinoma (ccRCC) cells proliferation and metastasis via altering of miR-508-3p/CXCL13 signal. Onco Targets Ther 2020;13:6051-62. [PMID: 32821115 DOI: 10.2147/OTT.S251436]
- Huang R, Zhang Y, Han B, Bai Y, Zhou R, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy 2017;13:1722-41. [PMID: 28786753 DOI: 10.1080/15548627.2017.1356975]
- Ji X, Shan L, Shen P, He M. Circular RNA circ_001621 promotes osteosarcoma cells proliferation and migration by sponging miR-578 and regulating VEGF expression. Cell Death Dis 2020;11:18. [PMID: 31907361 DOI: 10.1038/s41419-019-2204-y]
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. [PMID: 31395983 DOI: 10.1038/s41576-019-0158-7]
- Li S, Wang Q. Hsa_circ_0081534 increases the proliferation and invasion of nasopharyngeal carcinoma cells through regulating the miR-508-5p/FN1 axis. Aging (Albany NY) 2020;12:20645-57. [PMID: 33082297 DOI: 10.18632/aging.103963]
- Li Y, Gu J, Xu F, Zhu Q, Chen Y, et al. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Brief Bioinform 2021;22:bbaa225. [PMID: 33003204 DOI: 10.1093/bib/bbaa225]
- Liu CX, Chen LL. Circular RNAs: characterization, cellular roles, and applications. Cell 2022;185:2016-34. [PMID: 35584701 DOI: 10.1016/j.cell.2022.04.021]
- Liu S, Deng X, Zhang J. Identification of dysregulated serum miR-508-3p and miR-885-5p as potential diagnostic biomarkers of clear cell renal carcinoma. Mol Med Rep 2019;20:5075-83. [PMID: 31661117 DOI: 10.3892/mmr.2019.10762]
- Liu Y, Ma C, Qin X, Yu H, Shen L, et al. Circular RNA circ_001350 regulates glioma cell proliferation, apoptosis, and metastatic properties by acting as a miRNA sponge. J Cell Biochem 2019;120:15280-87. [PMID: 31020693 DOI: 10.1002/jcb.28795]
- Meng S, Zhou H, Feng Z, Xu Z, Tang Y, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017;16:94. [PMID: 28535767 DOI: 10.1186/s12943-017-0663-2]
- Mondal S, Adhikari N, Banerjee S, Amin SA, Jha T. Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: a minireview. Eur J Med Chem 2020;194:112260. [PMID: 32224379 DOI: 10.1016/j.ejmech.2020.112260]
- Nielsen AF, Bindereif A, Bozzoni I, Hanan M, Hansen TB, et al. Best practice standards for circular RNA research. Nat Methods 2022;19:1208-20. [PMID: 35618955 DOI: 10.1038/s41592-022-01487-2]
- Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol 2014;16:896-913. [PMID: 24842956 DOI: 10.1093/neuonc/nou087]
- Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J 2019;38:e100836. [PMID: 31343080 DOI: 10.15252/embj.2018100836]
- Rong D, Sun H, Li Z, Liu S, Dong C, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 2017;8:73271-81. [PMID: 29069868 DOI: 10.18632/oncotarget.19154]
- Singh R, Ha SE, Wei L, Jin B, Zogg H, et al. miR-10b-5p rescues diabetes and gastrointestinal dysmotility. Gastroenterology 2021;160:1662-78.e18. [PMID: 33421511 DOI: 10.1053/j.gastro.2020.12.062]
- Sun J, Zhao B, Du K, Liu P. TRAF6 correlated to invasion and poor prognosis of glioblastoma via elevating MMP9 expression. Neuroreport 2019;30:127-33. [PMID: 30571666 DOI: 10.1097/WNR.0000000000001171]
- Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci 2014;15:9331-42. [PMID: 24865493 DOI: 10.3390/ijms15069331]
- Wang J, Zhang ZQ, Li FQ, Chen JN, Gong X, et al. Triptolide interrupts rRNA synthesis and induces the RPL23-MDM2-p53 pathway to repress lung cancer cells. Oncol Rep 2020;43:1863-74. [PMID: 32236588 DOI: 10.3892/or.2020.7569]
- Wu R, Zeng J, Yuan J, Deng X, Huang Y, et al. MicroRNA-210 overexpression promotes psoriasis-like inflammation by inducing Th1 and Th17 cell differentiation. J Clin Invest 2018;128:2551-68. [PMID: 29757188 DOI: 10.1172/JCI97426]
- Xin J, Zhang XY, Sun DK, Tian LQ, Xu P. Up-regulated circular RNA hsa_circ_0067934 contributes to glioblastoma progression through activating PI3K-AKT pathway. Eur Rev Med Pharmacol Sci 2019;23:3447-54. [PMID: 31081099 DOI: 10.26355/eurrev_201904_17709]
- Xue B, Chuang C-H, Prosser HM, Fuziwara CS, Chan C, et al. miR-200 deficiency promotes lung cancer metastasis by activating Notch signaling in cancer-associated fibroblasts. Genes Dev 2021;35:1109-22. [PMID: 34301766 DOI: 10.1101/gad.347344.120]
- Yang M, Zhou Y, Deng H, Zhou H, Cheng S, et al. Ribosomal protein L23 drives the metastasis of hepatocellular carcinoma via upregulating MMP9. Front Oncol 2021;11:779748. [PMID: 34926291 DOI: 10.3389/fonc.2021.779748]
- Yang Y, Gao X, Zhang M, Yan S, Sun C, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst 2018;110:304-15. [PMID: 28903484 DOI: 10.1093/jnci/djx166]
- Yoshida K, Yokoi A, Yamamoto Y, Kajiyama H. ChrXq27.3 miRNA cluster functions in cancer development. J Exp Clin Cancer Res 2021;40:112. [PMID: 33766100 DOI: 10.1186/s13046-021-01910-0]
- Zhang SJ, Chen X, Li CP, Li XM, Liu C, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci 2017;58:6500-9. [PMID: 29288268 DOI: 10.1167/iovs.17-22698]
- Zhao L, Wang W, Xu L, Yi T, Zhao X, et al. Integrative network biology analysis identifies miR-508-3p as the determinant for the mesenchymal identity and a strong prognostic biomarker of ovarian cancer. Oncogene 2019;38:2305-19. [PMID: 30478449 DOI: 10.1038/s41388-018-0577-5]
- Zhao L, Guo Y, Guo Y, Ji X, Fan D, et al. Effect and mechanism of circRNAs in tumor angiogenesis and clinical application. Int J Cancer 2022;150:1223-32. [PMID: 34724210 DOI: 10.1002/ijc.33863]
- Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer 2020;19:172. [PMID: 33317550 DOI: 10.1186/s12943-020-01286-3]
- Zhou X, Liu K, Cui J, Xiong J, Wu H, et al. Circ-MBOAT2 knockdown represses tumor progression and glutamine catabolism by miR-433-3p/GOT1 axis in pancreatic cancer. J Exp Clin Cancer Res 2021;40:124. [PMID: 33832516 DOI: 10.1186/s13046-021-01894-x]