Recent advances in genetically modified large-animal models of human diseases
1Medical Research Center, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou, Guangdong, China
2Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, Guangdong, China
3Center for Reproductive Genetics and Reproductive Medicine, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, Guangdong, China
4Guangzhou Laboratory, Guangzhou, Guangdong, China
*Correspondence to: Chunwei Cao, E-mail: caochw5@mail.sysu.edu.cn
Received: May 23 2022; Revised: July 27 2022; Accepted: August 1 2022; Published Online: November 29 2022
Cite this paper:
Jing Zhang, Xiaoyue Sun and Chunwei Cao. Recent advances in genetically modified large-animal models of human diseases. BIO Integration 2022; 3(4): 161–171.
DOI: 10.15212/bioi-2022-0018. Available at: https://bio-integration.org/
Download citation
© 2022 The Authors. This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See https://bio-integration.org/copyright-and-permissions/
Abstract
Large-animal models show greater advantages than rodents in recapitulating human genetic diseases, primarily because of their higher similarity to humans in terms of anatomy, physiology and genetics. Notably, as genome-editing technologies have rapidly improved, particularly transcription activator-like effector nuclease (TALEN) and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 (CRISPR-associated protein 9) systems, their application in biomedical research has accelerated. A variety of genetically modified large-animal models, including non-human primates, pigs, dogs, bovines and sheep, have been produced to recapitulate human inherited disorders, thus providing novel biological and translational insights. Here, we review recent progress in the generation of large-animal models over the past 5 years and summarize their use in studying human genetic diseases, focusing on the nervous system, cardiovascular and metabolic systems, the immune system, xenotransplantation, the reproductive system and embryonic development.
Keywords
CRISPR/Cas9, human inherited diseases, large animal models, TALEN, translational medicine.
Introduction
Animal models, which are essential in biological and medical research, greatly promote advances in genetic research on human diseases. Among them, large-animal models have advantages because of their greater similarity to humans in terms of anatomy and physiology (Table S1) [1]. In addition, large-animal models show higher heterogeneity in genetic backgrounds, thus mirroring the genetic diversity of humans, and are genetically closer to humans than rodent models. Non-human primates (NHPs) are the model most closely resembling humans in evolution, genetics, physiology, the aging process, behavioral symptoms and pathological changes. Other large-animal models, including pigs, dogs, bovines and sheep, have been extensively studied to mimic human genetic diseases. However, their wide use in studies has been limited by a lack of efficient genetic engineering tools in these large animals. In recent years, new gene-editing technologies, mainly TALEN [2], CRISPR [3] and base editing [4], have made rapid progress and provided highly efficient tools for developing genetically modified large-animal models. Large-animal models are increasingly used to study genetic diseases, owing to their high efficiency and simplicity and the flexibility of CRISPR/Cas9 systems [5]. To date, two conventional pipelines have been established for the generation of genetically modified large-animal models [6]. One method is based on somatic-cell nuclear transfer (SCNT) combined with a genome-editing system [7]. The other involves generating gene-modified animals in a single step via microinjection of the CRISPR/Cas9 genome-editing system into zygotes, without a need for gene editing of somatic cells in vitro [8]. The ability to introduce genes or variants into animal genomes has allowed for mechanistic investigation into the genetic contributions of specific genes to human diseases. Here, we review the recent development of large-animal models and their applications in human genetic diseases over the past 5 years, focusing on disorders associated with the nervous, cardiovascular and metabolic, immune and reproductive systems.
Nervous system diseases
Nervous system diseases are a group of disorders associated with impairment of the central and peripheral nervous system. Research attention has focused on neurodegenerative disorders, which are accompanied by several representative symptoms in patients, including extrapyramidal and pyramidal movement disorders, as well as cognitive or behavioral disorders [9]. Human diseases associated with neurodegenerative disorders mainly include Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD) and infantile neuronal ceroid lipofuscinosis (CLN1), each of which has specific clinical symptoms and etiologies. In PD, studies have shown that both genetic and environmental factors are associated with pathogenesis. Patients with PD mainly present with muscle stiffness, tremors, unsteady gait and balance and coordination difficulties [10]. AD is the most common type of dementia affecting the older population, and patients with AD present a progressive decline in cognition, memory and behavioral skills, which is probably driven by β-amyloid protein deposition and intracellular accumulation of hyperphosphorylated tau protein [11]. HD is a rare inherited neurodegenerative disorder caused by a trinucleotide repeat (CAG) expansion in the HTT gene. Patients with HD exhibit uncontrolled choreatic movements, behavioral and psychiatric problems, and dementia [12]. CLN1 is a rare genetic disease caused by genetic changes in the PPT1 gene, which contribute to a deficiency in the soluble lysosomal enzyme palmitoyl protein thioesterase-1 (PPT1). Clinically, patients with CLN1 show severe neuronal degeneration, cortical thinning and overall brain atrophy [13]. Importantly, neurodegenerative diseases are age dependent [14], and previous studies have shown that mouse PD models do not fully replicate human pathological manifestations, probably because rodents lack major anatomical features found in humans, such as distinguishable subdivisions of the globus pallidus and a subthalamic nucleus [15, 16]. Therefore, the establishment of large-animal models for reproducing neurodegenerative lesions has become an attractive choice. Recently, Li et al. have built the first rhesus monkey model of etiological PD by co-editing the PINK1 and DJ-1 genes in the substantia nigra region of the monkey brain with the CRISPR/Cas9 system delivered by adeno-associated virus serotype 9 (AAV9). This transgenic monkey model simulates PD phenotypes well, such as bradykinesia, tremor and postural instability, which are accompanied by the key pathological characteristics of PD, including severe loss of nigral dopaminergic neurons and the presence of α-synuclein pathology within the gene-edited substantia nigra [17]. In 2019, Yang et al. generated a CRISPR/Cas9-mediated PINK1-deleted monkey model and observed robust early-onset neurodegeneration in various brain regions; this model should provide important information for studying the function of PINK1 and progressive neurodegeneration [18]. In addition, this group has also demonstrated that PINK1 kinase activity rather than its mitochondrial function is a selective requirement for neuronal survival in primate brains, and have further suggested that PINK1 kinase dysfunction might be associated with human PD and other pathological conditions [19]. Furthermore, to create an ideal inherited PD minipig model, Zhu et al. have produced Bama minipig models by introducing three PD-causing missense variants in the SCNA gene (E46K, H50Q and G51D) by using CRISPR/Cas9-mediated gene editing combined with SCNT. Owing to the absence of α-synuclein-immunopositive pathology at 3 months of age, these pig models still must be developed to investigate the presence of PD-like pathological features [20]. In contrast to PD, AD is associated with clinical memory and cognition deficits as well as pathologically neurofibrillary tangles and amyloid plaques [21]. Recently, an AD transgenic pig model bearing mutations in hAPP (K670N/M671L, I716V and V717I), hTau (P301L) and hPS1 (M146V and L286P) has been developed. This model shows high expression of target genes in tissues, particularly in the brain, and exhibits hallmarks of damaged neurons consisting of Aβ-40/42, total Tau and GFAP; therefore, it may serve as an ideal model for studying AD pathogenesis [22]. A knock-in (KI) pig model carrying the mutant huntingtin (HTT) gene shows consistent movement and behavioral abnormalities, which are accompanied by striking and selective degeneration of striatal medium spiny neurons [15]. This work first demonstrated that the overt and selective neurodegeneration seen in patients with HD can be reproduced by endogenously expressed mutant proteins in large mammals. Additionally, SURF1−/− pig models have been generated with TALENs and CRISPRs to recapitulate Leigh syndrome associated with cytochrome c oxidase (COX) deficiency. SURF−/− pigs show failure to thrive, a short life span and muscle weakness; in newborn piglets, delayed central nervous system development is observed in the absence of clear COX deficiency [23]. A CLN1 sheep model with CRISPR/Cas9-mediated insertion of human PPT1 (R151X) has been successfully constructed, which exhibits behavioral and motor deficits as well as hallmarks of brain atrophy, thus providing substantial opportunities for further revealing the mechanisms and discovering a potential treatment for this form of neurodegenerative disease [24].
Moreover, several large-animal models have recently been created for modeling other nervous system diseases, such as autism spectrum disorder (ASD), Rett syndrome (RTT) and tuberous sclerosis (TSC). In 2019, Zhou et al. reported that SHANK-mutant monkeys exhibit sleep disturbances, motor deficits and repetitive behaviors, as well as social and learning impairments, which resemble characteristics of ASD and Phelan–McDermid syndrome [25]. Likewise, Tu et al. have developed a cynomolgus monkey model with SHANK3 gene disruption, which exhibits the core disease phenotypes of ASD. Furthermore, their results have indicated that treatment with the antidepressant fluoxetine alleviates the abnormal behaviors and brain activity, thus indicating the advantages of using NHPs for ASD modeling [26, 27]. In addition, Qin et al., concentrating on nonsyndromic ASD, have found that specific knockout of giant ANK2 in monkeys does not generate nonsyndromic ASD-like behaviors, but gives rise to pronounced brain structural alterations [28]. Of interest, TALEN-edited MECP2 cynomolgus monkey models show major phenotypic similarities to human patients with RTT, thus suggesting that MECP2 gene-edited mutant monkeys will be valuable for dissecting disease pathogenesis and developing potential therapeutic strategies for RTT [29]. Moreover, Tph2 knockout (KO) pig models have been generated and provided important insights into behavioral abnormalities induced by 5-HT deficiency [30]. In addition, a recent study has developed a pig model by introducing a monoallelic mutation in the TSC1 gene with the CRISPR system. TSC1+/- pigs develop the clinical features observed in patients with TSC, including cardiac rhabdomyoma and subependymal nodules, which are absent in mouse TSC models [31]. STXBP1 is essential for neurotransmitter release, and STXBP1 (R292H)-mutated monkeys created through base editing show core symptoms of STXBP1 encephalopathy, thus providing a suitable animal model for STXBP1 encephalopathy [32]. Furthermore, Beraldi et al. have established an ATM−/− pig model for modeling ataxia telangiectasia. Interestingly, ATM−/− pigs not only simulate the neurological phenotype but also show other pathological features of patients with ataxia telangiectasia, including altered thymus structure, dysregulation of the immune system and sterility [33].
Cardiovascular and metabolic diseases
Cardiovascular diseases refer to a group of disorders affecting the heart and blood vessels [34]. Metabolic diseases are conditions in which abnormal metabolic processes occur, primarily including dyslipidemia and perturbation of amino acid metabolism [35]. In studies of inherited cardiovascular and metabolic diseases, pigs have received the greatest attention among large-animal models. Notably, pigs present many advantages over other animals, such as similar cardiovascular anatomy and cardiac physiology to those in humans [36], as well as a similar heart size to that in humans [37]. Therefore, they provide an ideal model for cardiovascular disease research. For example, pigs and humans express β-MHC in ventricles, which play important roles in regulating heart rates and maintaining the cardiac output. However, fast α-MHC, instead of β-MHC, has been found to be expressed in mouse ventricles—a characteristic notably different from those in humans and pigs [38]. In 2021, Gabriel et al. generated a CRISPR-edited pig model with SAP130 mutation for modeling human congenital heart disease, which is rare in pigs. This pig model, which manifests coronary heart disease with tricuspid dysplasia and tricuspid atresia associated with early embryonic lethality, provides opportunities for research in surgical operation and testing ventricular assist devices [39]. In addition, Montag et al., using the TALEN system, have successfully constructed an MYH7 (R723G)-mutant pig model mimicking human familial hypertrophic cardiomyopathy [40]. A TALEN-induced SGCD KO pig mimicking human genetic cardiomyopathy has been generated and found to exhibit symptoms of systolic dysfunction, myocardial tissue degeneration and sudden death, thus potentially enabling the development of preclinical therapies [41]. Chen et al. have found that transgenic pF9 KO pigs carrying the human coagulation factor IX show partial amelioration of bleeding; this model may be used to explore the pathological process of hemophilia [42]. Moreover, Zhang et al. have produced DUOX2D409G/D409G mutant pigs through ENU mutagenesis and demonstrated that the TR-KLF9 axis is responsible for the blood cell development in hypothyroidism [43].
Furthermore, many groups have chosen pigs or dogs for modeling human metabolic diseases. Studies have shown that, compared with those in mice, the lipoprotein profiles and metabolism patterns of pigs are overall more similar to those in humans [44]. However, a large proportion of cholesterol transport is mediated by high-density lipoprotein in mice—an aspect clearly different from the low-density-lipoprotein delivery in humans and pigs [45]. Therefore, several groups have produced pig mutants via the CRISPR/Cas9 system for modeling human diabetes by targeting the INS [46], NGN3 [47] and hIAPP [48] genes, whose functions are associated with pancreatic development. Furthermore, Wang et al. first generated permanent neonatal diabetes mellitus (PNDM) dog models carrying GCK point mutations by using the BE3 system. These models exhibit similar features to those in patients with GCK-PNDM and thus may serve as ideal animal models to study this disease [49]. Moreover, ASGR1-deficient [50], ApoE KO [51] and ApoE/LDLR dKO [52] pigs have been found to be ideal models for human cardiovascular diseases associated with lipid metabolism, particularly high cholesterol. Likewise, ApoE KO dog models produced with CRISPR/Cas9 also show advanced severe hypercholesterolemia and atherosclerosis characterized by stenosis and occlusion of arteries, together with stroke and gangrene [53]. These pig and dog models will be invaluable in developing and evaluating new therapies, including endovascular procedures, to treat atherosclerosis and related disorders. Yao et al. have found that OSBPL2 KO pigs, generated through a combination of CRISPR/Cas9 and SCNT techniques, show hypercholesterolemia and progressive hearing loss, thus confirming the roles of OSBPL2 gene in nonsyndromic hearing loss and providing opportunities to unravel the potential relationships between auditory dysfunction and dyslipidemia/hypercholesterolemia [54]. Yin et al. have found that MC3R KO pigs, exhibiting increased body weight and body fat, can be used to reveal the physiological roles of MC3R in energy homeostasis [55]. In addition, chronic inflammation has been demonstrated to contribute to obesity and metabolic diseases, particularly metaflammation [56]. Zhang et al. have generated triple transgenic pigs through CRISPR/Cas9-mediated KI of GIPRdn, hIAPP and PNPLA3I148M and found that the model develops metabolic disorders accompanied by inflammation activation; thus, this model may be ideal for investigating metabolic inflammation [57]. Interestingly, Zheng et al., in adipose-specific UCP1 KI pigs, have uncovered crucial roles of UCP1 in protecting the cardiovascular system through inhibiting tissue inflammatory responses [58].
Using FAH−/− pigs created by CRISPR/Cas9, an ideal animal model of hereditary tyrosinemia type 1 (HT1), Gu et al. have found that, before intrauterine death, direct intracytoplasmic delivery of CRISPR-Cas9 targeting the HPD gene reprograms the tyrosine metabolism pathway and protects pigs against FAH-deficiency-induced lethal liver injury, thus providing a therapeutic option for the treatment of HT1 [59]. Additionally, in 2020, Koppes et al. successfully produced a CRISPR/Cas9-mediated PAH-null pig model recapitulating human phenylalanine hydroxylase–deficient phenylketonuria (PKU) and enabling investigation of therapeutic interventions [60]. In addition, in 2021, Kaiser et al. generated a PAHhR408W/hR408W PKU pig model by using a TALEN system and found that this model mimics human phenotypes and responds well to dietary phenylalanine restriction [61]. Importantly, these pig models for human PKU have introduced perspectives in the development of therapeutic interventions and have unique value in gene-therapy studies.
Immune system diseases
Previous studies have indicated that rodent models have shortcomings in immunology research [62], and the differences in immune responses between rodents and humans might be attributable to genetics, lifespan, living environment and specific species-pathogen relationships [63]. Large-animal models can offer unique biological advantages in understanding human immunology and may be able to address questions that rodent models cannot answer [64]. Remarkably, in contrast with rodents, large animals show greater similarities to humans in terms of immune system development and response, including immune cell development, innate immunity, regional immunity and infectious immunity. The pig immune system has been demonstrated to resemble that in humans for more than 80% of parameters, whereas the mouse immune system has similarity for approximately 10% of parameters [65]. Swine models have been widely used in studies of autoimmune and immune-mediated inflammatory diseases. Interestingly, transgenic pigs with leptin overexpression show symptoms of systemic lupus erythematosus, including anemia, leukopenia and thrombocytopenia, along with kidney and liver impairment. However, glucocorticoid therapies have been found to partially relieve the autoimmune symptoms. The leptin transgenic pig model is valuable for investigating the roles of adipocytokines in the modulation of immune responses [66]. Zhang et al. have successfully established complement protein C3 KO pigs, which can be used to delineate the roles of C3 in pathology and physiology [67]. In addition, Li et al. have found that pigs carrying NLRP3 (R259W) homozygous mutations mimic aspects of human cryopyrin-associated periodic syndrome, such as early mortality, poor growth and spontaneous systemic inflammation symptoms [68]. Song et al. have built a pig model of human familial acne inversa, an inflammatory skin condition, by introducing an NCSTN+/R117X heterozygous point mutation, and have further elucidated the mechanism underlying the development of this condition [69]. Large-animal models are also increasingly being developed and used in studies on infectious diseases. Yugo et al. have successfully established JH−/− gnotobiotic pigs with knockout of the immunoglobulin heavy chain. Compared with wild-type pigs, JH−/− pigs show lower levels of HEV replication and enlarged livers after HEV infection, thus suggesting that JH−/− pigs may provide an efficient animal model to mimic HEV-specific lesions and dissect the mechanisms of HEV pathogenesis [70]. Notably, the emergence of the novel virus SARS-CoV-2 has greatly affected human life worldwide. ACE2, the major entry receptor for this virus, acts on the kinin-kallikrein, renin-angiotensin and coagulation systems, which have been implicated in the pathogenesis of severe cases of the related disease, COVID-19 [71]. To reproduce severe cases of COVID-19, Du et al. have successfully established hACE2 KI pigs, and have detected higher expression of hACE2 protein in the lungs, kidneys, testes and intestines, similarly to the conditions observed in humans [72].
Xenotransplantation
Xenotransplantation research in large-animal models has made major advances. The current research focus in this field includes immune rejection, physiological incompatibilities and the risk of microbial transmission in conducting transplantation [73]. Pigs have received substantial attention, owing to their similarities with humans in terms of biological features. The enzyme 1,3-galactosyltransferase (Galα (1,3) Gal), encoded by GGTA1, acts as a key factor in xenograft rejection, by catalyzing the synthesis of αGal. In 2017, TALEN modified CMAHKO/GTKO/sh-TNFRI-Fc/hHO-1 quadruply modified pigs have been found to overcome ultra-acute and acute anti-inflammatory rejection of xenografts [74]. In addition, Adams et al. have found that the kidneys transplanted from CRISPR/Cas9-mediated double-xenoantigen Gal-Sda KO pigs into chemical immunosuppression rhesus monkeys show prolonged xenogeneic survival times as long as 435 days; they have also revealed that early graft rejection is mediated by IgM antibody, but the 435-day graft loss might nonetheless have resulted from IgG-antibody-mediated rejection [75]. In addition, Kim et al. have performed renal transplantation from αGal KO/CD55 transgenic pigs into rhesus macaques, and have found that early xenograft rejection was induced by abundant CD4+ cell infiltration, and later rejection is mediated by antibodies [76]. In addition, Fu et al. have demonstrated that skin grafts from GGTA1−/−β2M−/−CIITA−/− triple knockout (GBC-3KO) pigs show significantly prolonged survival in mice, thus indicating that GBC-3KO effectively decreases xenogeneic immune responses [77]. Rao et al. have developed HLA-G1+/GGTA1 KO pigs through transgenic expression of HLA-G1+ in GGTA1 KO pigs, and have demonstrated that these donors suppress the activation and proliferation of monkey and human T, B and natural killer (NK) cells [78]. In 2019, Watanabe et al. transplanted pig lung xenografts expressing human-CD47 (hCD47) into baboons and found that the xenografts had a prolonged survival time for 8 weeks [79]. In addition, two reported cases of pig-to-human kidney xenotransplantation have shown that genetically modified kidney xenografts from GGTA1−/− pigs are viable and can function in brain-dead human recipients for 54 hours, without signs of hyperacute rejection [80]. In regard to the xeno-organ overgrowth problem, Hinrichs et al. have eliminated the growth hormone receptor in GGTA1 KO/hCD46+/hTHBD+ pigs and found that GHR knockout decreases the intrinsic growth potential of pig xeno-organs [81].
In addition to being an organ-transplant donor, immunodeficient pigs are also favorable recipient research models. Choi et al. have reported that RAG2−/− pigs representing severe combined immunodeficiency (SCID) show advantages over Rag2−/− SCID mice, because the mice show intense, infrequent and mild clusters of CD3+, CD4+ and CD8+ signals. The gene expression of T, B or NK cell maturation in RAG2−/− SCID pigs is less than that in Rag2−/− SCID mice [82]. Nelson et al. have found that RAG2−/−FAH−/− immunodeficient pigs can receive infusions of human liver cells, although the NK cells are a barrier to the expansion of hepatocytes [83]. Furthermore, Ren et al. have demonstrated that IL2RG−/Y pigs allow for the development of human melanoma-derived tumors and thus may serve as hosts for human cancer [84]. In addition, Hendricks-Wenger et al. have found that RAG2/IL2RG double KO pigs permit growth of human pancreatic adenocarcinomas, whose electrical properties and responses to irreversible electroporation are similar to those of excised human pancreatic cancer tumors, thus suggesting a key step in the development of immune humanized SCID pig models [85]. Dong et al. have successfully generated porcine endogenous-retrovirus-inactivated pigs by using CRISPR/Cas9, thus addressing the safety concerns in clinical xenotransplantation [86].
Reproductive system and embryonic development
Large-animal models also have advantages over rodent models in human reproduction studies. For example, large animals provide valuable resources for investigating folliculogenesis, which is challenging in rodents. Shi et al. have generated bone morphogenesis protein 15 (BMP15) KO pigs with the CRISPR/Cas9 system and found that this model shows an infertility phenotype. In detail, BMP15 depletion obstructs follicle development at preantral stages, thus resulting in the development of many structurally abnormal follicles and consequently leading to streaky ovaries and lack of a pronounced estrus cycle [87]. In addition, large animals are ideal models for studies on mammalian embryonic development. Recently, two groups have found that SRY KO pigs generated by CRISPR/Cas9 [6] and SRY KI bovines generated by TALEN [88] show sex reversal, thus further demonstrating the conserved roles of SRY in mammalian sex determination and differentiation. Zhou et al. have constructed a pig parthenogenetic embryo model by targeting the PDHA1 gene with CRISPR/Cas9 at the four-cell stage and found that early embryonic development is blocked, and histone acetylation significantly decreases, thereby demonstrating the critical roles of PADH1 in zygotic genome activation in porcine embryos [89]. Kilian et al. have constructed OCT4 KO bovine embryos by using CRISPR and SCNT; their results have indicated that, as in human early embryonic development, OCT4 is necessary for maintaining NANOG-positive epiblast cells in the inner cell mass of bovine blastocysts, in contrast to findings in mice [90]. Likewise, Daigneault et al. have shown that disruption of POU5F1 in bovine embryos by the CRISPR/Cas9 system contributes to embryonic arrest at the morula stage, thereby preventing blastocyst formation. Moreover, conservation of POU5F1 functions in embryonic development has been observed in bovines and humans, in contrast to mice [91]. Thus, these studies highlight that bovine embryogenesis provides an outstanding model for understanding human early development.
Other diseases
In 2019, Dorado et al. established a Yucatan minipig model of Hutchinson-Gilford progeria syndrome through a heterozygous LMNA c.1824 C>T mutation. This model shows severe growth retardation, lipodystrophy, skin and bone alterations, cardiovascular disease and death around puberty, in agreement with symptoms in humans [92]. Furthermore, in 2020, monkey models bearing LMNA c.1824 C>T, created by base editing, were also found to show the typical symptoms of Hutchinson-Gilford progeria syndrome [93]. To investigate the biological function of longevity protein SIRT6 in primates, Zhang et al. have generated SIRT6-null cynomolgus monkey models with the CRISPR/Cas9 system. The KO monkeys died shortly after birth and exhibited severe prenatal developmental retardation, thus mimicking human perinatal lethality syndrome [94]. In addition, Tsukiyama et al. have introduced mutations in PKD1 and produced a cynomolgus macaque model of autosomal dominant polycystic kidney disease. PKD1 depletion in heterozygous monkeys leads to severe cyst formation, primarily in the collecting ducts, and cyst formation perinatally in distal tubules, thus somewhat reflecting the initial pathology in humans [95].
Duchenne muscular dystrophy (DMD) is a common hereditary childhood myopathy caused by DMD gene mutations [96]. Notably, dogs spontaneously produce DMD [97]. Multiple canine DMD models have also been established with genome-editing technologies, thus providing opportunities for in vivo gene-therapy trials. In 2018, Amoasii et al. performed in vivo CRISPR gene editing in a deltaE50-MD dog model of DMD by using adeno-associated viruses (AAVs); this gene-editing treatment restores the dystrophin in skeletal muscle and cardiac muscle [98]. Furthermore, Moretti et al. have demonstrated that intramuscular injection of AAV9-Cas9-gE51 in a deltaE50-MD swine model induces expression of a shortened dystrophin (DMDΔ51–52), thereby improving skeletal muscle function [99]. In addition, Li et al. have successfully established FSI-I-I KI pigs by using CRISPR/Cas9. The myofiber sizes in FSI-I-I KI pigs were significantly greater than those in wild-type pigs, thus indicating great promise for treatment of human muscular dystrophy [100]. However, although dystrophin expression was restored, Hakim et al. have found that AAV-CRISPR treatment leads to a Cas9-specific immune response in multiple dystrophic canine models, thus posing major challenges for CRISPR gene-editing therapies [101].
In 2019, Gao et al. used CRISPR/Cas9 and SCNT to knock out the HR gene in pigs; piglets with mutations exhibit a lack of hair on the eyelids, and abnormalities in the thymus and peripheral blood [102]. Furthermore, genome editing of the FGF5 [103], EDAR [104] and VEGF [105] genes in goats and sheep has been found to significantly increase hair growth and hair-follicle density, thus suggesting potential roles of these genes in follicle diseases. Han et al. have produced HOXC13 KO pigs with CRISPR/Cas9. This model shows a diminished number of follicles and disarray in hair follicle cables, thus mimicking human ectodermal dysplasia-9 [106]. In addition, a pig model bearing the deep intronic mutation IVS49-727 A>G in ABCA12 shows hyperkeratotic skin and a response to systemic retinoid treatment, thereby recapitulating human harlequin ichthyosis [107]. In 2018, Li et al. successfully generated pigs carrying the TWIST2 E75K mutation and TYR Q68Stop variant by using BE3 and SCNT; the phenotypes were consistent with those of human ablepharon macrostomia syndrome and oculocutaneous albinism type 1 (OCA1), respectively [108]. Moreover, Zhang et al. have established a COL2A1 KO pig model exhibiting severe skeletal dysplasia and tracheal collapse, to investigate the pathogenesis of early skeletal developmental defects [109]. Likewise, Williams et al. have created a sheep hypophosphatasia model through CRISPR/Cas9-mediated KI of an ALPL gene mutation (1077 C>G). The KI sheep exhibit diminished serum alkaline phosphatase activity, tail vertebral bone size and metaphyseal faring, thus providing a unique platform for bone research [110]. Additionally, Watanabe et al. have created a SALL1-null pig model displaying a nephrogenic phenotype, thus potentially offering a nephrogenic niche for human kidney regeneration [111]. In 2018, FGFR2-IIIb was found to play a role in lung branching morphogenesis in pigs overexpressing dominant-negative FGFR2-IIIb made by SCNT [112]. Hai et al. have produced a pig model carrying the c.740 T>C (L247S) mutation in the MITF gene for modeling human Waardenburg syndrome type 2A [113]. The group has further performed CRISPR-Cas9-mediated gene therapy to correct phenotypes including anophthalmia and hearing loss [114]. Moreover, Engevik et al. have generated a pig model by introducing the MYO5B P663L mutation with TALENs; the pigs mimic human microvilli inclusion body disease [115].
Conclusions and future prospects
In conclusion, large-animal models have been widely used to mimic human genetic diseases, particularly disorders of the nervous system, cardiovascular and metabolic systems, immune system, reproductive system and embryonic development. Since 2017, more than 80 research articles regarding the construction of large-animal models for human diseases have been published (in NHPs, pigs, dogs, cattle, sheep and goats, on the basis of PubMed searches). The development of genome-editing tools has greatly revolutionized the field, thus making genetic engineering and genome editing of large-animal genomes simpler, and more precise and efficient. Moreover, large-animal models provide major advantages in modeling specific human diseases that rodent models may fail to faithfully recapitulate. Importantly, large-animal models may also provide unique or unexpected insights for better understanding of human diseases.
The increasing number of large-animal models has heralded a new phase of understanding of the complex conditions of human inherited disease. These advances have further offered prospects for therapeutic applications of gene therapy in large animals. Many large animals, such as pigs [116], dogs [117] and NHPs [118], have been used for the assessment of gene-transfer techniques or gene-therapy treatment trials. Notably, owing to their relatively longer lifespan and size, large animals have substantial advantages in addressing concerns regarding the long-term efficacy and safety of gene-therapy approaches. Large-animal models also present a unique translational framework for validating and testing novel therapeutic tools, such as new genome-editing or gene-delivery systems. For example, in 2021, Musunuru et al. delivered a CRISPR base-editing system by using lipid nanoparticles and generated durable low-density-lipoprotein cholesterol for 8 months in monkey livers, thus providing a promising strategy to target in vivo treatment for liver diseases [119]. Currently, omics technologies, including genomics, transcriptomics, proteomics and metabolomics, have been used extensively in biomedical studies and yielded valuable new findings. Similarly, increasing amounts of omics data have been obtained in large-animal models; these data may be used to choose suitable animal models according to different scenarios. Specially, recent technological advances have enabled omics investigations of single cells or restricted spatial areas, thus revealing information on gene expression within individual cells and also capturing spatial gene expression profiles. Prominently, four studies have built a single-cell atlas including 33 human organs [120–123]. Han et al. completed a mouse cell atlas including more than 40 mouse organs and tissues [124]. Recently, an adult monkey cell atlas covering 45 different tissues has been created [125]. Furthermore, the BodyMap transcriptome containing approximately 31 adult pig tissues has been reported [126]. In addition, single-cell RNA sequencing of various tissues or organs derived from dog lung immune cell populations [127], sheep germ cells [128] and bovine sperm cells [129] have been obtained. Theoretically, comparison of these omics data between human and animal models should provide systemic information for estimating the biological relevance or similarity of the model to humans, thus helping researchers select the right model according to a broad perspective.
In 1990, the first gene-therapy trial for a rare inherited disease known as SCID was initiated [130]. Owing to great advances in genome-editing and gene-delivery systems, gene therapy has become a major research field that holds great promise for treating human genetic diseases. Importantly, gene therapy has been clinically used in human genetic disorders, such as thalassemia, DMD, cystic fibrosis, eye disorders, metabolic disorders and blood coagulation disorders. Trials of gene therapy for infectious diseases, including acquired immunodeficiency syndrome and COVID-19, have also been reported recently [131]. In fact, large-animal models have substantial advantages in gene-therapy studies. Similarly to humans, large animals commonly have a heterogeneous genetic background, unlike inbred mice; have long lifespans enabling investigation of long-term effects; and have relevant organs and body sizes matching those of neonates or children, thus providing unique opportunities to address issues associated with gene therapy [132]. Specially, large animals have been considered suitable for modeling human neurodegenerative diseases, largely because large animals have similar brain sizes to those in humans [133] and have a sulcated cortex, which is not observed in rodents [134]. Because of the advantages of their long life span, gene therapy targeting cystic fibrosis in large animals has been successfully demonstrated [135]. Large animals have also been used for developing and assessing novel gene-delivery techniques. For example, the clinically well established and catheter-based antegrade delivery methods, as evaluated in large-animal models, are safer than other invasive delivery techniques, which is a critical aspect for treating patients with cardiac disease [136]. Furthermore, numerous gene-delivery systems, including viral and non-viral gene-delivery systems, have recently been developed for use in gene therapy. Indeed, large animals are ideal models for evaluating the specificity, safety and efficiency of these gene-delivery systems.
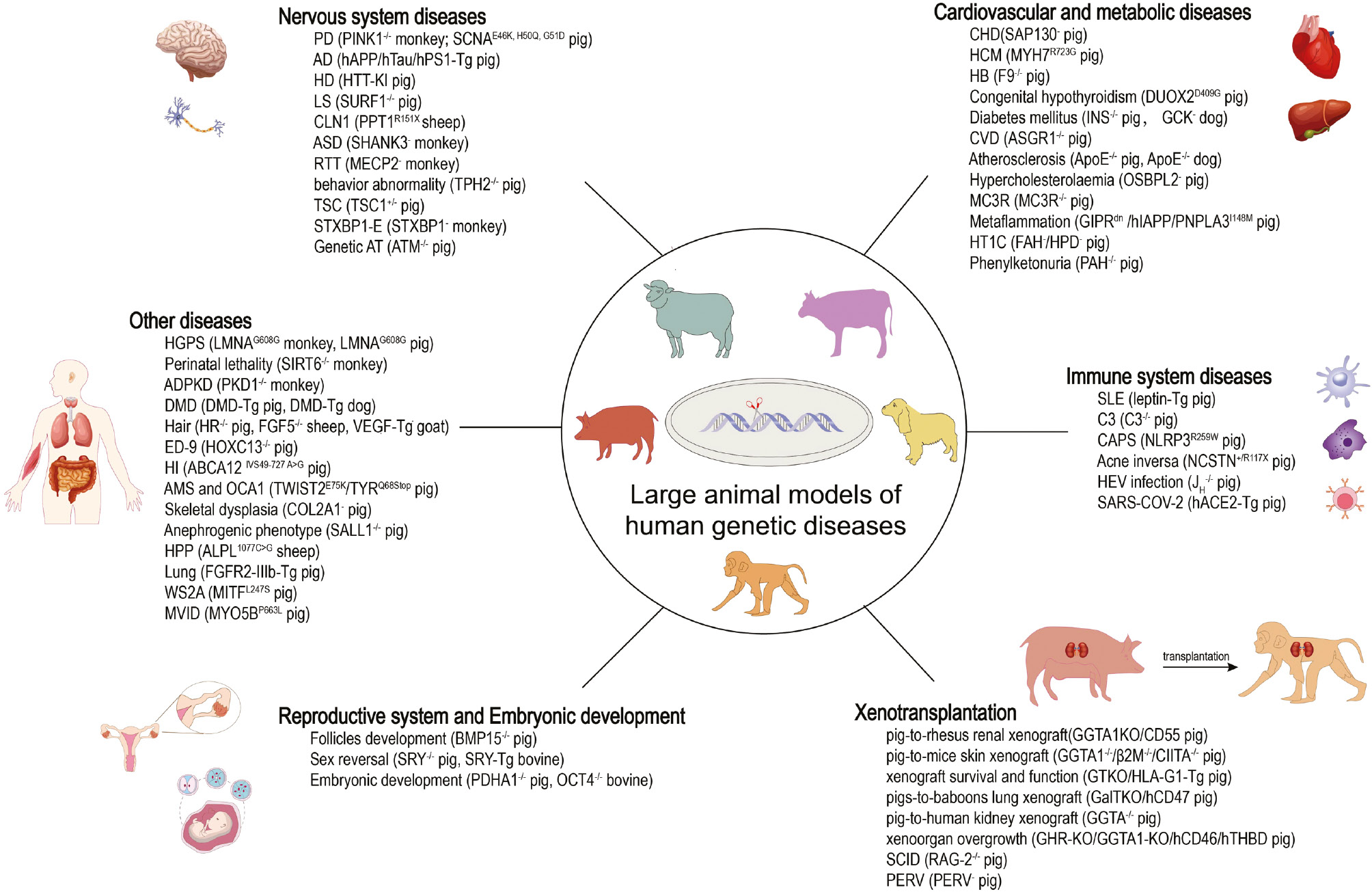
Collectively, the production of large-animal models for human inherited diseases has made considerable advances resulting from the rapid progress in genome editing in large animals (Figure 1; Table S2). Further development of genetic manipulation tools with higher gene modification efficiency, as well as better gene-delivery efficiency and specificity, will support broader application of large-animal models in translational biomedical research.
Figure 1 The genetically modified large animal models of human diseases. −/− represents konckout. Tg means transgenic. – means mutation.
Supplementary Table
Click here to download Supplementary Table.
Acknowledgements
This study was supported by the National Nature Science Foundation of China (grant no. 32170612).
References
- Barre-Sinoussi F, Montagutelli X. Animal models are essential to biological research: issues and perspectives. Future Sci OA 2015;1:FSO63. [PMID: 28031915 DOI: 10.4155/fso.15.63]
- Joung JK, Sander JD. INNOVATION TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 2013;14:49-55. [PMID: 23169466 DOI: 10.1038/nrm3486]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281-308. [PMID: 24157548 DOI: 10.1038/nprot.2013.143]
- Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 2018;19:770-8. [PMID: 30323312 DOI: 10.1038/s41576-018-0059-1]
- Wei C, Liu J, Yu Z, Zhang B, Gao G, et al. TALEN or Cas9 – rapid, efficient and specific choices for genome modifications. J Genet Genomics 2013;40:281-9. [PMID: 23790627 DOI: 10.1016/j.jgg.2013.03.013]
- Kurtz S, Lucas-Hahn A, Schlegelberger B, Gohring G, Niemann H, et al. Knockout of the HMG domain of the porcine SRY gene causes sex reversal in gene-edited pigs. P Natl Acad Sci USA 2021;118:e2008743118. [PMID: 33443157 DOI: 10.1073/pnas.2008743118]
- Wilmut I, Bai Y, Taylor J. Somatic cell nuclear transfer: origins, the present position and future opportunities. Philos Trans R Soc Lond B Biol Sci 2015;370:20140366. [DOI: 10.1098/rstb.2014.0366]
- Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 2014;24:372-5. [PMID: 24481528 DOI: 10.1038/cr.2014.11]
- Dugger BN, Dickson DW. Pathology of neurodegenerative diseases. Cold Spring Harb Perspect Biol 2017;9:a028035. [PMID: 28062563 DOI: 10.1101/cshperspect.a028035]
- Bartels AL, Leenders KL. Parkinson’s disease: the syndrome, the pathogenesis and pathophysiology. Cortex 2009;45:915-21. [PMID: 19095226 DOI: 10.1016/j.cortex.2008.11.010]
- Lamptey RNL, Chaulagain B, Trivedi R, Gothwal A, Layek B, et al. A review of the common neurodegenerative disorders: current therapeutic approaches and the potential role of nanotherapeutics. Int J Mol Sci 2022;23:1851. [PMID: 35163773 DOI: 10.3390/ijms23031851]
- McAllister B, Gusella JF, Landwehrmeyer GB, Lee JM, MacDonald ME, et al. Timing and impact of psychiatric, cognitive, and motor abnormalities in huntington disease. Neurology 2021;96:e2395-e406. [PMID: 33766994 DOI: 10.1212/WNL.0000000000011893]
- Hawkins-Salsbury JA, Cooper JD, Sands MS. Pathogenesis and therapies for infantile neuronal ceroid lipofuscinosis (infantile CLN1 disease). Biochim Biophys Acta 2013;1832:1906-9. [PMID: 23747979 DOI: 10.1016/j.bbadis.2013.05.026]
- Heemels MT. Neurodegenerative diseases. Nature 2016;539:179. [PMID: 27830810 DOI: 10.1038/539179a]
- Yan S, Tu ZC, Liu ZM, Fan NN, Yang HM, et al. A huntingtin knockin pig model recapitulates features of selective neurodegeneration in huntington’s disease. Cell 2018;173:989-1002. [PMID: 29606351 DOI: 10.1016/j.cell.2018.03.005]
- Morton AJ, Avanzo L. Executive decision-making in the domestic sheep. Plos One 2011;6:e15752. [PMID: 21305061 DOI: 10.1371/journal.pone.0015752]
- Li H, Wu S, Ma X, Li X, Cheng T, et al. Co-editing PINK1 and DJ-1 genes via adeno-associated virus-delivered CRISPR/Cas9 system in adult monkey brain elicits classical parkinsonian phenotype. Neurosci Bull 2021;37:1271-88. [PMID: 34165772 DOI: 10.1007/s12264-021-00732-6]
- Yang WL, Li SH, Li XJ. A CRISPR monkey model unravels a unique function of PINK1 in primate brains. Mol Neurodegener 2019;14:17. [DOI: 10.1186/s13024-019-0321-9]
- Yang WL, Guo XY, Tu ZC, Chen XS, Han R, et al. PINK1 kinase dysfunction triggers neurodegeneration in the primate brain without impacting mitochondrial homeostasis. Protein Cell 2022;13:26-46. [PMID: 34800266 DOI: 10.1007/s13238-021-00888-x]
- Zhu XX, Zhong YZ, Ge YW, Lu KH, Lu SS. CRISPR/Cas9-mediated generation of guangxi bama minipigs harboring three mutations in alpha-synuclein causing parkinson’s disease. Sci Rep 2018;8:12420. [PMID: 30127453 DOI: 10.1038/s41598-018-30436-3]
- Karch CM, Cruchaga C, Goate AM. Alzheimer’s disease genetics: from the bench to the clinic. Neuron 2014;83:11-26. [PMID: 24991952 DOI: 10.1016/j.neuron.2014.05.041]
- Lee SE, Hyun H, Park R, Choi Y, Son YJ, et al. Production of transgenic pig as an Alzheimer’s disease model using a multi-cistronic vector system. Plos One 2017;12:e0177933. [PMID: 28586343 DOI: 10.1371/journal.pone.0177933]
- Quadalti C, Brunetti D, Lagutina I, Duchi R, Perota A, et al. SURF1 knockout cloned pigs: Early onset of a severe lethal phenotype. BBA-Mol Basis Dis 2018;1864:2131-42. [PMID: 29601977 DOI: 10.1016/j.bbadis.2018.03.021]
- Eaton SL, Proudfoot C, Lillico SG, Skehe P, Kline RA, et al. CRISPR/Cas9 mediated generation of an ovine model for infantile neuronal ceroid lipofuscinosis (CLN1 disease). Sci Rep 2019;9:9891. [PMID: 31289301 DOI: 10.1038/s41598-019-45859-9]
- Zhou Y, Sharma J, Ke Q, Landman R, Yuan JL, et al. Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature 2019;570:326-31. [DOI: 10.1038/s41586-019-1278-0]
- Tu ZC, Zhao H, Li B, Yan S, Wang L, et al. CRISPR/Cas9-mediated disruption of SHANK3 in monkey leads to drug-treatable autism-like symptoms. Hum Mol Genet 2019;28:561-71. [PMID: 30329048 DOI: 10.1093/hmg/ddy367]
- Zhao H, Tu Z, Xu H, Yan S, Yan H, et al. Altered neurogenesis and disrupted expression of synaptic proteins in prefrontal cortex of SHANK3-deficient non-human primate. Cell Res 2017;27(10):1293-7. [DOI: 10.1038/cr.2017.95]
- Qin DD, Zhou JK, He XC, Shen XY, Li C, et al. Depletion of giant ANK2 in monkeys causes drastic brain volume loss. Cell Discov 2021;7:113. [PMID: 34845196 DOI: 10.1038/s41421-021-00336-4]
- Chen YC, Yu JH, Niu YY, Qin DD, Liu HL, et al. Modeling rett syndrome using TALEN-Edited MECP2 Mutant Cynomolgus Monkeys. Cell 2017;169:945-55. [PMID: 28525759 DOI: 10.1016/j.cell.2017.04.035]
- Li Z, Yang HY, Wang Y, Zhang ML, Liu XR, et al. Generation of tryptophan hydroxylase 2 gene knockout pigs by CRISPR/Cas9-mediated gene targeting. J Biomed Res 2017;31:445-52. [PMID: 28866660 DOI: 10.7555/JBR.31.20170026]
- Li X, Hu T, Liu J, Fang B, Geng X, et al. A Bama miniature pig model of monoallelic TSC1 mutation for human tuberous sclerosis complex. J Genet Genomics 2020;47:735-42. [PMID: 33612456 DOI: 10.1016/j.jgg.2020.11.005]
- Lu Z, He S, Jiang J, Zhuang L, Wang Y, et al. Base-edited cynomolgus monkeys mimic core symptoms of STXBP1 encephalopathy. Mol Ther 2022;30:2163-75. [PMID: 35797994 DOI: 10.1016/j.ymthe.2022.06.020]
- Beraldi R, Meyerholz DK, Savinov A, Kovacs AD, Weimer JM, et al. Genetic ataxia telangiectasia porcine model phenocopies the multisystemic features of the human disease. Biochim Biophys Mol Basis Dis 2017;1863:2862-70. [PMID: 28746835 DOI: 10.1016/j.bbadis.2017.07.020]
- Farley A, McLafferty E, Hendry C. The cardiovascular system. Nurs Stand 2012;27:35-9. [PMID: 23240514 DOI: 10.7748/ns2012.10.27.9.35.c9383]
- Scholl-Burgi S, Sass JO, Zschocke J, Karall D. Amino acid metabolism in patients with propionic acidaemia. J Inherit Metab Dis 2012;35:65-70. [PMID: 21113738 DOI: 10.1007/s10545-010-9245-9]
- Lillywhite HB, Zippel KC, Farrell AP. Resting and maximal heart rates in ectothermic vertebrates. Comp Biochem Phys A Mol Integr Physicol 1999;124:369-82. [PMID: 10682235 DOI: 10.1016/s1095-6433(99)00129-4]
- Cui J, Li J, Mathison M, Tondato F, Mulkey SP, et al. A clinically relevant large-animal model for evaluation of tissue-engineered cardiac surgical patch materials. Cardiovasc Revasc Med 2005;6:113-20. [PMID: 16275607 DOI: 10.1016/j.carrev.2005.07.006]
- Milani-Nejad N, Janssen PM. Small and large animal models in cardiac contraction research: advantages and disadvantages. Pharmacol Ther 2014;141:235-49. [PMID: 24140081 DOI: 10.1016/j.pharmthera.2013.10.007]
- Gabriel GC, Devine W, Redel BK, Whitworth KM, Samuel M, et al. Cardiovascular development and congenital heart disease modeling in the pig. J Am Heart Assoc 2021;10:e021631. [PMID: 34219463 DOI: 10.1161/JAHA.121.021631]
- Montag J, Petersen B, Flogel AK, Becker E, Lucas-Hahn A, et al. Successful knock-in of Hypertrophic Cardiomyopathy-mutation R723G into the MYH7 gene mimics HCM pathology in pigs. Sci Rep 2018;8:4786. [PMID: 29555974 DOI: 10.1038/s41598-018-22936-z]
- Matsunari H, Honda M, Watanabe M, Fukushima S, Suzuki K, et al. Pigs with delta-sarcoglycan deficiency exhibit traits of genetic cardiomyopathy. Lab Invest 2020;100:887-99. [PMID: 32060408 DOI: 10.1038/s41374-020-0406-7]
- Chen JH, An BY, Yu B, Peng XH, Yuan HM, et al. CRISPR/Cas9-mediated knockin of human factor IX into swine factor IX locus effectively alleviates bleeding in hemophilia B pigs. Haematologica 2021;106:829-37. [PMID: 31974191 DOI: 10.3324/haematol.2019.224063]
- Zhang Y, Xue Y, Cao C, Huang J, Hong Q, et al. Thyroid hormone regulates hematopoiesis via the TR-KLF9 axis. Blood 2017;130:2161-70. [PMID: 28972010 DOI: 10.1182/blood-2017-05-783043]
- Kaabia Z, Poirier J, Moughaizel M, Aguesse A, Billon-Crossouard S, et al. Plasma lipidomic analysis reveals strong similarities between lipid fingerprints in human, hamster and mouse compared to other animal species. Sci Rep 2018;8:15893. [DOI: 10.1038/s41598-018-34329-3]
- Lee-Rueckert M, Escola-Gil JC, Kovanen PT. HDL functionality in reverse cholesterol transport–Challenges in translating data emerging from mouse models to human disease. Biochim Biophys Acta 2016;1861:566-83. [PMID: 26968096 DOI: 10.1016/j.bbalip.2016.03.004]
- Cho B, Kim SJ, Lee EJ, Ahn SM, Lee JS, et al. Generation of insulin-deficient piglets by disrupting INS gene using CRISPR/Cas9 system. Transgenic Res 2018;27:289-300. [PMID: 29691708 DOI: 10.1007/s11248-018-0074-1]
- Sheets TP, Park KE, Park CH, Swift SM, Powell A, et al. Targeted Mutation of NGN3 Gene Disrupts Pancreatic Endocrine Cell Development in Pigs. Sci Rep 2018;8:3582. [PMID: 29483633 DOI: 10.1038/s41598-018-22050-0]
- Zou XD, Ouyang HS, Yu TT, Chen X, Pang DX, et al. Preparation of a new type 2 diabetic miniature pig model via the CRISPR/Cas9 system. Cell Death Dis 2019;10:823. [DOI: 10.1038/s41419-019-2056-5]
- Wang XM, Liang YH, Zhao JP, Li Y, Gou SX, et al. Generation of permanent neonatal diabetes mellitus dogs with glucokinase point mutations through base editing. Cell Discov 2021;7:92. [DOI: 10.1038/s41421-021-00304-y]
- Xie B, Shi X, Li Y, Xia B, Zhou J, et al. Deficiency of ASGR1 in pigs recapitulates reduced risk factor for cardiovascular disease in humans. PLoS Genet 2021;17(11):e1009891. [PMID: 34762653 DOI: 10.1371/journal.pgen.1009891]
- Fang B, Ren XY, Wang Y, Li Z, Zhao LH, et al. Apolipoprotein E deficiency accelerates atherosclerosis development in miniature pigs. Dis Model Mech 2018;11:dmm036632. [PMID: 30305304 DOI: 10.1242/dmm.036632]
- Huang L, Hua ZD, Xiao HW, Cheng Y, Xu K, et al. CRISPR/Cas9-mediated ApoE(-/-) and LDLR-/- double gene knockout in pigs elevates serum LDL-C and TC levels. Oncotarget 2017;8:37751-60. [PMID: 28465483 DOI: 10.18632/oncotarget.17154]
- Zhao H, Zhao JP, Wu D, Sun ZL, Hua Y, et al. Dogs lacking Apolipoprotein E show advanced atherosclerosis leading to apparent clinical complications. Sci China Life Sci 2021;65:1342-56. [PMID: 34705220 DOI: 10.1007/s11427-021-2006-y]
- Yao J, Zeng HS, Zhang M, Wei QJ, Wang Y, et al. OSBPL2-disrupted pigs recapitulate dual features of human hearing loss and hypercholesterolaemia. J Genet Genomics 2019;46:379-87. [PMID: 31451425 DOI: 10.1016/j.jgg.2019.06.006]
- Yin YJ, Hao HY, Xu XB, Shen LC, Wu WJ, et al. Generation of an MC3R knock-out pig by CRSPR/Cas9 combined with somatic cell nuclear transfer (SCNT) technology. Lipids Health Dis 2019;18:122. [DOI: 10.1186/s12944-019-1073-9]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature 2006;444:860-7. [PMID: 17167474 DOI: 10.1038/nature05485]
- Zhang K, Tao C, Xu J, Ruan J, Xia J, et al. CD8(+) T cells involved in metabolic inflammation in visceral adipose tissue and liver of transgenic pigs. Front Immunol 2021;12:690069. [PMID: 34322121 DOI: 10.3389/fimmu.2021.690069]
- Gu P, Hui X, Zheng Q, Gao Y, Jin L, et al. Mitochondrial uncoupling protein 1 antagonizes atherosclerosis by blocking NLRP3 inflammasome-dependent interleukin-1beta production. Sci Adv 2021;7:eabl4024. [PMID: 34878840 DOI: 10.1126/sciadv.abl4024]
- Gu P, Yang Q, Chen BZ, Bie YN, Liu W, et al. Genetically blocking HPD via CRISPR-Cas9 protects against lethal liver injury in a pig model of tyrosinemia type I. Mol Ther-Meth Clin D 2021;21:530-47. [PMID: 33997102]
- Koppes EA, Redel BK, Johnson MA, Skvorak KJ, Ghaloul-Gonzalez L, et al. A porcine model of phenylketonuria generated by CRISPR/Cas9 genome editing. Jci Insight 2020;5:e141523. PMID: 33055427 DOI: 10.1172/jci.insight.141523]
- Kaiser RA, Carlson DF, Allen KL, Webster DA, VanLith CJ, et al. Development of a porcine model of phenylketonuria with a humanized R408W mutation for gene editing. PLoS One 2021;16:e0245831. [PMID: 33493163 DOI: 10.1371/journal.pone.0245831]
- Liu SX, Du YC, Zeng T. A mini-review of the rodent models for alcoholic liver disease: shortcomings, application, and future prospects. Toxicol Res (Camb) 2021;10:523-30. [PMID: 34141166 DOI: 10.1093/toxres/tfab042]
- Masopust D, Sivula CP, Jameson SC. Of mice, dirty mice, and men: using mice To understand human immunology. J Immunol 2017;199:383-8. [PMID: 28696328 DOI: 10.4049/jimmunol.1700453]
- Hein WR, Griebel PJ. A road less travelled: large animal models in immunological research. Nat Rev Immunol 2003;3:79-84. [PMID: 12511878 DOI: 10.1038/nri977]
- Dawson H. A comparative assessment of the pig, mouse and human genomes. In: McAnulty PA, Dayan AD, Ganderup NC, Hastings KL, editors. The minipig in biomedical research. Boca Raton, FL, USA: CRC Press; 2011. pp. 323342.
- Chen JC, Zeng WQ, Pan WR, Peng C, Zhang JL, et al. Symptoms of systemic lupus erythematosus are diagnosed in leptin transgenic pigs. PLoS Biol 2018;16:e2005354. [PMID: 30169503 DOI: 10.1371/journal.pbio.2005354]
- Zhang W, Wang G, Wang Y, Jin Y, Zhao LH, et al. Generation of complement protein C3 deficient pigs by CRISPR/Cas9-mediated gene targeting. Sci Rep 2017;7:5009 . [PMID: 28694465 DOI: 10.1038/s41598-017-05400-2]
- Li W, Shi L, Zhuang Z, Wu H, Lian M, et al. Engineered pigs carrying a gain-of-function NLRP3 homozygous mutation can survive to adulthood and accurately recapitulate human systemic spontaneous inflammatory responses (vol 205, pg 2532, 2020). J Immunol 2020;205:2532-44. [PMID: 32958688 DOI: 10.4049/jimmunol.1901468]
- Song R, Liu K, Wang Y, Qin G, Xiao M, et al. A pig model carrying heterozygous point mutation of NCSTN simulates familial acne inversa and reveals dysregulated cholesterol biosynthesis via the Notch-pAMPK-HMGCR pathway. Sci Bull 2021;66:2343-6. [DOI: 10.1016/j.scib.2021.05.022]
- Yugo DM, Heffron CL, Ryu J, Uh K, Subramaniam S, et al. Infection dynamics of hepatitis E Virus in wild-type and immunoglobulin heavy chain knockout JH (-/-) gnotobiotic piglets. J Virol 2018;92:e01208-18. [PMID: 30111571 DOI: 10.1128/JVI.01208-18]
- Sidarta-Oliveira D, Jara CP, Ferruzzi AJ, Skaf MS, Velander WH, et al. SARS-CoV-2 receptor is co-expressed with elements of the kinin-kallikrein, renin-angiotensin and coagulation systems in alveolar cells. Sci Rep 2020;10:19522. [PMID: 33177594 DOI: 10.1038/s41598-020-76488-2]
- Du XG, Guo ZH, Fan WH, Hai T, Gao F, et al. Establishment of a humanized swine model for COVID-19. Cell Discov 2021;7:70. [PMID: 34404772 DOI: 10.1038/s41421-021-00313-x]
- Denner J. Xenotransplantation-Progress and Problems: A Review. J Transpl Technol Res 2014;04 . [DOI: 10.4172/2161-0991.1000133]
- Kim GA, Lee EM, Jin JX, Lee S, Taweechaipaisankul A, et al. Generation of CMAHKO/GTKO/shTNFRI-Fc/HO-1 quadruple gene modified pigs. Transgenic Res 2017;26:435-45. [PMID: 28553699 DOI: 10.1007/s11248-017-0021-6]
- Adams AB, Kim SC, Martens GR, Ladowski JM, Estrada JL, et al. Xenoantigen deletion and chemical immunosuppression can prolong renal xenograft survival. Ann Surg 2018;268:564-73. [PMID: 30048323 DOI: 10.1097/SLA.0000000000002977]
- Kim SC, Mathews DV, Breeden CP, Higginbotham LB, Ladowski J, et al. Long-term survival of pig-to-rhesus macaque renal xenografts is dependent on CD4 T cell depletion. Am J Transplant 2019;19:2174-85. [PMID: 30821922 DOI: 10.1111/ajt.15329]
- Fu R, Fang M, Xu K, Ren J, Zou J, et al. Generation of GGTA1-/-beta2M-/-CIITA-/- pigs using CRISPR/Cas9 technology to alleviate xenogeneic immune reactions. Transplantation 2020;104:1566-73. [PMID: 32732833 DOI: 10.1097/TP.0000000000003205]
- Rao JS, Hosny N, Kumbha R, Naqvi RA, Singh A, et al. HLA-G1(+) expression in GGTA1KO pigs suppresses human and monkey anti-pig T, B and NK cell responses. Front Immunol 2021;12:730545. [PMID: 34566993 DOI: 10.3389/fimmu.2021.730545]
- Watanabe H, Ariyoshi Y, Pomposelli T, Takeuchi K, Ekanayake-Alper DK, et al. Intra-bone bone marrow transplantation from hCD47 transgenic pigs to baboons prolongs chimerism to >60 days and promotes increased porcine lung transplant survival. Xenotransplantation 2020;27:e12552. [PMID: 31544995 DOI: 10.1111/xen.12552]
- Montgomery RA, Stern JM, Lonze BE, Tatapudi VS, Mangiola M, et al. Results of two cases of pig-to-human kidney Xenotransplantation. N Engl J Med 2022;386:1889-98. [PMID: 35584156 DOI: 10.1056/NEJMoa2120238]
- Hinrichs A, Riedel EO, Klymiuk N, Blutke A, Kemter E, et al. Growth hormone receptor knockout to reduce the size of donor pigs for preclinical xenotransplantation studies. Xenotransplantation 2021;28:e12664 . [PMID: 33241624 DOI: 10.1111/xen.12664]
- Choi YJ, Kim E, Reza A, Hong K, Song H, et al. Recombination activating gene-2(null) severe combined immunodeficient pigs and mice engraft human induced pluripotent stem cells differently. Oncotarget 2017;8:69398-407. [PMID: 29050212 DOI: 10.18632/oncotarget.20626]
- Nelson ED, Larson E, Joo DJ, Mao SN, Glorioso J, et al. Limited expansion of human hepatocytes in FAH/RAG2-deficient Swine. Tissue Eng Pt A 2022;28:150-60. [PMID: 34309416 DOI: 10.1089/ten.TEA.2021.0057]
- Ren J, Yu D, Fu R, An P, Sun R, et al. IL2RG-deficient minipigs generated via CRISPR/Cas9 technology support the growth of human melanoma-derived tumours. Cell Proliferat 2020;53:e12863. [PMID: 32871045 DOI: 10.1111/cpr.12863]
- Hendricks-Wenger A, Aycock KN, Nagai-Singer MA, Coutermarsh-Ott S, Lorenzo MF, et al. Establishing an immunocompromised porcine model of human cancer for novel therapy development with pancreatic adenocarcinoma and irreversible electroporation. Sci Rep 2021;11:7584. [DOI: 10.1038/s41598-021-87228-5]
- Niu D, Wei HJ, Lin L, George H, Wang T, et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 2017;357:1303-7. [PMID: 28798043 DOI: 10.1126/science.aan4187]
- Shi X, Tang T, Lin Q, Liu H, Qin Y, et al. Efficient generation of bone morphogenetic protein 15-edited Yorkshire pigs using CRISPR/Cas9. Biol Reprod 2020;103:1054-68. [PMID: 32761111 DOI: 10.1093/biolre/ioaa138]
- Wang M, Sun Z, Ding F, Wang H, Li L, et al. Efficient TALEN-mediated gene knockin at the bovine Y chromosome and generation of a sex-reversal bovine. Cell Mol Life Sci 2021;78:5415-25. [PMID: 34047803 DOI: 10.1007/s00018-021-03855-1]
- Zhou WJ, Niu YJ, Nie ZW, Kim JY, Xu YN, et al. Nuclear accumulation of pyruvate dehydrogenase alpha 1 promotes histone acetylation and is essential for zygotic genome activation in porcine embryos. BBA-Mol Cell Res 2020;1867:118648. [DOI: 10.1016/j.bbamcr.2020.118648]
- Simmet K, Zakhartchenko V, Philippou-Massier J, Blum H, Klymiuk N. OCT4/POU5F1 is required for NANOG expression in bovine blastocysts. P Natl Acad Sci USA 2018;115:2770-5. [PMID: 29483258 DOI: 10.1073/pnas.1718833115]
- Daigneault BW, Rajput S, Smith GW, Ross PJ. Embryonic POU5F1 is Required for Expanded Bovine Blastocyst Formation. Sci Rep 2018;8:7753. [DOI: 10.1038/s41598-018-25964-x]
- Dorado B, Ploen GG, Barettino A, Macias A, Gonzalo P, et al. Generation and characterization of a novel knockin minipig model of Hutchinson-Gilford progeria syndrome. Cell Discov 2019;5:16. [DOI: 10.1038/s41421-019-0084-z]
- Wang F, Zhang W, Yang Q, Kang Y, Fan Y, et al. Generation of a Hutchinson-Gilford progeria syndrome monkey model by base editing. Protein Cell 2020;11:809-24. [PMID: 32729022 DOI: 10.1007/s13238-020-00740-8]
- Zhang W, Wan H, Feng G, Qu J, Wang J, et al. SIRT6 deficiency results in developmental retardation cynomolgus monkeys. Nature 2018;560:661-5. [PMID: 30135584 DOI: 10.1038/s41586-018-0437-z]
- Tsukiyama T, Kobayashi K, Nakaya M, Iwatani C, Seita Y, et al. Monkeys mutant for PKD1 recapitulate human autosomal dominant polycystic kidney disease. Nat Commun 2019;10:5517. [DOI: 10.1038/s41467-019-13398-6]
- Huang Y, Yu S, Wu Z, Tang B. Genetics of hereditary neurological disorders in children. Transl Pediatr 2014;3:108-19. [PMID: 26835329 DOI: 10.3978/j.issn.2224-4336.2014.03.04]
- Walmsley GL, Arechavala-Gomeza V, Fernandez-Fuente M, Burke MM, Nagel N, et al. A duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient cavalier king charles spaniels is amenable to exon 51 skipping. PLoS One 2010;5:e8647. [PMID: 20072625 DOI: 10.1371/journal.pone.0008647]
- Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 2018;362:86-90. [PMID: 30166439 DOI: 10.1126/science.aau1549]
- Moretti A, Fonteyne L, Giesert F, Hoppmann P, Meier AB, et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med 2020;26:207-14. [PMID: 31988462 DOI: 10.1038/s41591-019-0738-2]
- Li M, Tang X, You W, Wang Y, Chen Y, et al. HMEJ-mediated site-specific integration of a myostatin inhibitor increases skeletal muscle mass in porcine. Mol Ther Nucleic Acids 2021;26:49-62. [PMID: 34513293 DOI: 10.1016/j.omtn.2021.06.011]
- Hakim CH, Kumar SRP, Perez-Lopez DO, Wasala NB, Zhang D, et al. Cas9-specific immune responses compromise local and systemic AAV CRISPR therapy in multiple dystrophic canine models. Nat Commun. 2021;12:6769 . [DOI: 10.1038/s41467-021-26830-7]
- Gao QS, Xuan MF, Luo ZB, Paek HJ, Kang JD, et al. Hairless-knockout piglets generated using the clustered regularly interspaced short palindromic repeat/CRISPR-associated-9 exhibit abnormalities in the skin and thymus. Exp Anim 2019;68:519-29. [PMID: 31308290 DOI: 10.1538/expanim.19-0018]
- Zhang R, Li Y, Jia K, Xu X, Li Y, et al. Crosstalk between androgen and Wnt/beta-catenin leads to changes of wool density in FGF5-knockout sheep. Cell Death Dis 2020;11:407. [PMID: 32472005 DOI: 10.1038/s41419-020-2622-x]
- Hao F, Yan W, Li XC, Wang H, Wang YM, et al. Generation of cashmere goats carrying an EDAR gene mutant using CRISPR-Cas9-mediated genome editing. Int J Biol Sci 2018;14:427-36. [PMID: 29725264 DOI: 10.7150/ijbs.23890]
- Hu X, Hao F, Li XC, Xun ZY, Gao Y, et al. Generation of VEGF knock-in Cashmere goat via the CRISPR/Cas9 system. Int J Biol Sci 2021;17:1026-40. [PMID: 33867826 DOI: 10.7150/ijbs.55559]
- Han K, Liang LP, Li L, Ouyang Z, Zhao BT, et al. Generation of Hoxc13 knockout pigs recapitulates human ectodermal dysplasia-9. Hum Mol Genet 2017;26:184-91. [PMID: 28011715 DOI: 10.1093/hmg/ddw378]
- Wang X, Cao CW, Li YS, Hai T, Jia QT, et al. A harlequin ichthyosis pig model with a novel ABCA12 mutation can be rescued by acitretin treatment. J Mol Cell Biol 2019;11:1029-41. [PMID: 30925591 DOI: 10.1093/jmcb/mjz021]
- Li ZF, Duan XY, An XM, Feng T, Li P, et al. Efficient RNA-guided base editing for disease modeling in pigs. Cell Discov 2018;4:64. [PMID: 30588328 DOI: 10.1038/s41421-018-0065-7]
- Zhang BY, Wang CY, Zhang Y, Jiang Y, Qin YG, et al. A CRISPR-engineered swine model of COL2A1 deficiency recapitulates altered early skeletal developmental defects in humans. Bone 2020;137:115450. [PMID: 32450343 DOI: 10.1016/j.bone.2020.115450]
- Williams DK, Pinzon C, Huggins S, Pryor JH, Falck A, et al. Genetic engineering a large animal model of human hypophosphatasia in sheep. Sci Rep 2018;8:16945. [PMID: 30446691 DOI: 10.1038/s41598-018-35079-y]
- Watanabe M, Nakano K, Uchikura A, Matsunari H, Yashima S, et al. Anephrogenic phenotype induced by SALL1 gene knockout in pigs. Sci Rep 2019;9:8016. [PMID: 31142767 DOI: 10.1038/s41598-019-44387-w]
- Chen Q, Fang B, Wang Y, Li C, Li XX, et al. Overexpressing dominant-negative FGFR2-IIIb impedes lung branching morphogenesis in pigs. J Genet Genomics 2018;45:147-54. [PMID: 29576506 DOI: 10.1016/j.jgg.2018.02.002]
- Hai T, Guo WW, Yao J, Cao CW, Luo AL, et al. Creation of miniature pig model of human Waardenburg syndrome type 2A by ENU mutagenesis. Hum Genet 2017;136:1463-75. [PMID: 29094203 DOI: 10.1007/s00439-017-1851-2]
- Yao J, Wang Y, Cao CW, Song RG, Bi DF, et al. CRISPR/Cas9-mediated correction of MITF homozygous point mutation in a Waardenburg syndrome 2A pig model. Mol Ther-Nucl Acids 2021;24:986-99. [PMID: 34094716 DOI: 10.1016/j.omtn.2021.04.009]
- Engevik AC, Coutts AW, Kaji I, Rodriguez P, Ongaratto F, et al. Editing myosin VB gene to create porcine model of microvillus inclusion disease, with microvillus-lined inclusions and alterations in sodium transporters. Gastroenterology 2020;158:2236-49 e9. [PMID: 32112796 DOI: 10.1053/j.gastro.2020.02.034]
- Gabisonia K, Prosdocimo G, Aquaro GD, Carlucci L, Zentilin L, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 2019;569:418-22. [PMID: 31068698 DOI: 10.1038/s41586-019-1191-6]
- Nguyen GN, Everett JK, Kafle S, Roche AM, Raymond HE, et al. A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells. Nat Biotechnol 2021;39:47-55. [PMID: 33199875 DOI: 10.1038/s41587-020-0741-7]
- Pignataro D, Sucunza D, Rico AJ, Dopeso-Reyes IG, Roda E, et al. Gene therapy approaches in the non-human primate model of Parkinson’s disease. J Neural Transm (Vienna) 2018;125:575-89. [PMID: 28130586 DOI: 10.1007/s00702-017-1681-3]
- Musunuru K, Chadwick AC, Mizoguchi T, Garcia SP, DeNizio JE, et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 2021;593:429-34. [PMID: 34012082 DOI: 10.1038/s41586-021-03534-y]
- Tabula Sapiens C, Jones RC, Karkanias J, Krasnow MA, Pisco AO, et al. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 2022;376:eabl4896. [PMID: 35549404 DOI: 10.1126/science.abl4896]
- Eraslan G, Drokhlyansky E, Anand S, Fiskin E, Subramanian A, et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science 2022;376:eabl4290. [PMID: 35549429 DOI: 10.1126/science.abl4290]
- Conde CD, Xu C, Jarvis LB, Rainbow DB, Wells SB, et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 2022;376:eabl5197. [PMID: 35549406 DOI: 10.1126/science.abl5197]
- Suo C, Dann E, Goh I, Jardine L, Kleshchevnikov V, et al. Mapping the developing human immune system across organs. Science 2022:eabo0510. [PMID: 35549310 DOI: 10.1126/science.abo0510]
- Han X, Wang R, Zhou Y, Fei L, Sun H, et al. Mapping the Mouse Cell Atlas by Microwell-seq. Cell 2018;172:1091-107 e17. [PMID: 29474909 DOI: 10.1016/j.cell.2018.02.001]
- Han L, Wei X, Liu C, Volpe G, Zhuang Z, et al. Cell transcriptomic atlas of the non-human primate Macaca fascicularis. Nature 2022;604:723-31. [PMID: 35418686 DOI: 10.1038/s41586-022-04587-3]
- Jin L, Tang Q, Hu S, Chen Z, Zhou X, et al. A pig BodyMap transcriptome reveals diverse tissue physiologies and evolutionary dynamics of transcription. Nat Commun 2021;12:3715. [PMID: 34140474 DOI: 10.1038/s41467-021-23560-8]
- Fastres A, Pirottin D, Fievez L, Marichal T, Desmet CJ, et al. Characterization of the bronchoalveolar lavage fluid by single cell gene expression analysis in healthy dogs: a promising technique. Front Immunol 2020;11:1707. [PMID: 32849601 DOI: 10.3389/fimmu.2020.01707]
- Yang H, Ma J, Wan Z, Wang Q, Wang Z, et al. Characterization of sheep spermatogenesis through single-cell RNA sequencing. Faseb J 2021;35:e21187. [PMID: 33197070 DOI: 10.1096/fj.202001035RRR]
- Capra E, Turri F, Lazzari B, Cremonesi P, Gliozzi TM, et al. Small RNA sequencing of cryopreserved semen from single bull revealed altered miRNAs and piRNAs expression between High- and Low-motile sperm populations. BMC Genomics 2017;18:14. [DOI: 10.1186/s12864-016-3394-7]
- Blaese RM, Culver KW, Miller AD, Carter CS, Fleisher T, et al. T lymphocyte-directed gene therapy for ADA- SCID: initial trial results after 4 years. Science 1995;270:475-80. [PMID: 7570001 DOI: 10.1126/science.270.5235.475]
- Arabi F, Mansouri V, Ahmadbeigi N. Gene therapy clinical trials, where do we go? An overview. Biomed Pharmacother 2022;153:113324. [PMID: 35779421 DOI: 10.1016/j.biopha.2022.113324]
- Casal M, Haskins M. Large animal models and gene therapy. Eur J Hum Genet 2006;14:266-72. [PMID: 16333317 DOI: 10.1038/sj.ejhg.5201535]
- Vite CH, McGowan JC, Niogi SN, Passini MA, Drobatz KJ, et al. Effective gene therapy for an inherited CNS disease in a large animal model. Ann Neurol 2005;57:355-64. [PMID: 15732095 DOI: 10.1002/ana.20392]
- Wolfe JH. Gene therapy in large animal models of human genetic diseases. Introduction. ILAR J 2009;50:107-11. [PMID: 19293455 DOI: 10.1093/ilar.50.2.107]
- Potash AE, Wallen TJ, Karp PH, Ernst S, Moninger TO, et al. Adenoviral gene transfer corrects the ion transport defect in the sinus epithelia of a porcine CF model. Mol Ther 2013;21:947-53. [PMID: 23511247 DOI: 10.1038/mt.2013.49]
- Watanabe S, Leonardson L, Hajjar RJ, Ishikawa K. Cardiac gene delivery in large animal models: antegrade techniques. Methods Mol Biol 2017;1521:227-35. [PMID: 27910053 DOI: 10.1007/978-1-4939-6588-5_16]