Single-Cell RNA-Sequencing Integration Analysis Revealed Immune Cell Heterogeneity in Five Human Autoimmune Diseases
Announcing a new article publication for BIO Integration journal. Autoimmune diseases are a group of diseases caused by abnormal immune responses to functional body parts. Single-cell RNA-sequencing (scRNA-seq) technology provides transcriptomic information at the single-cell resolution, thus offering a new way to study autoimmune diseases. Most single-cell RNA-seq studies, however, have often focused on one type of autoimmune disease.
scRNA-seq data was integrated from peripheral blood cells of five different autoimmune diseases (IgA nephropathy [IgAN], Kawasaki disease [KD], multiple sclerosis [MS], Sjogren’s syndrome [SS], and systemic lupus erythematosus [SLE]). Dimensionality clustering, cellular communication analysis, re-clustering analysis of monocytes, NK cell populations, differential gene expression analysis, and functional enrichment was performed for all immune cells in these data.
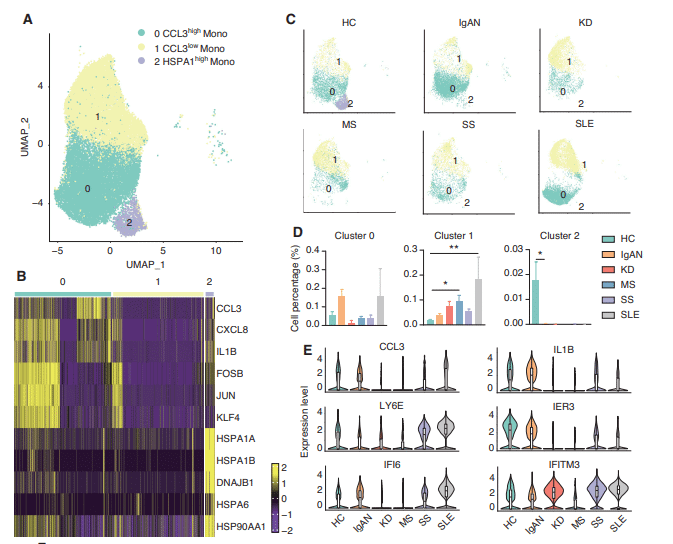
The scRNA-seq results of peripheral blood cells from five different autoimmune diseases (IgAN, KD, MS, SS, and SLE) were integrated. All samples contained 18 different immune cell subsets, although the cell cluster populations were different among the 5 diseases. Through intercellular communication network analysis, we determined that the signals of classical and non-classical monocytes were significantly enhanced in patients with IgAN and SLE. The signals of naïve B cells were increased in patients KD. Interestingly, the signals of NK and NK-T cells were enhanced in patients with SS, but reduced in patients with IgAN and SLE. Transcriptomic analysis of classical and non-classical monocyte subsets further revealed that pro-inflammatory cytokines and interferon-related genes, including CCL3, IL1B, ISG15, and IFI6, were specifically increased in patients with IgAN and SLE. Unlike monocytes, the number and NK marker genes were decreased in patients with IgAN and KD, but increased in patients with SS. Meanwhile, two NK-T cell subsets were exclusively found in SS.
In summary, based on an integration of the single-cell RNA-seq results, we demonstrated changes in the immune cell landscape of five different autoimmune diseases with respect to immune cell subsets, populations, differentially-expressed genes, and the cell-to-cell communication network. The data provides new insight to further explore the heterogeneity and similarity among different autoimmune diseases.
https://doi.org/10.15212/bioi-2023-0012
BIO Integration is fully open access journal which will allow for the rapid dissemination of multidisciplinary views driving the progress of modern medicine.
As part of its mandate to help bring interesting work and knowledge from around the world to a wider audience, BIOI will actively support authors through open access publishing and through waiving author fees in its first years. Also, publication support for authors whose first language is not English will be offered in areas such as manuscript development, English language editing and artwork assistance.
BIOI is now open for submissions; articles can be submitted online at:
https://mc04.manuscriptcentral.com/bioi
Please visit www.bio-integration.org to learn more about the journal.
Editorial Board: https://bio-integration.org/editorial-board/
Please visit www.bio-integration.org to learn more about the journal.
Editorial Board: https://bio-integration.org/editorial-board/
BIOI is available on the IngentaConnect platform (https://www.ingentaconnect.com/content/cscript/bioi) and at the BIO Integration website (www.bio-integration.org).
Submissions may be made using ScholarOne (https://mc04.manuscriptcentral.com/bioi).
There are no author submission or article processing fees.
Follow BIOI on Twitter @JournalBio; Facebook (https://www.facebook.com/BIO-Integration-Journal-108140854107716/) and LinkedIn (https://www.linkedin.com/company/bio-integration-journal/).
ISSN 2712-0074
eISSN 2712-0082
Single-Cell RNA-Sequencing Integration Analysis Revealed Immune Cell Heterogeneity in Five Human Autoimmune Diseases, Luo, Siweier; Wang, Le; Xiao, Yi; Cao, Chunwei; Liu, Qinghua; Zhou, Yiming; BIO Integration 2023, https://doi.org/10.15212/bioi-2023-0012