Aptamer-based Theranostics in Oncology: Design Strategies and Limitations
1Amity Institute of Pharmacy, Lucknow, Amity University Uttar Pradesh, Noida, Uttar Pradesh, India
2ERA College of Pharmacy, ERA University, Lucknow, Uttar Pradesh, India
*Correspondence to: Dr. Mohammad Yasir, Assistant Professor (G-III), Amity Institute of Pharmacy, Amity University Uttar Pradesh Lucknow Campus, Near Malhaur Railway Station, P.O. Chinhat, Lucknow 226028, Uttar Pradesh, India, Mobile: +919179417284. E-mail: yasirmohammad6@gmail.com; myasir@lko.amity.edu
Received: January 19 2024; Revised: February 21 2024; Accepted: March 26 2024; Published Online: April 10 2024
Cite this paper:
Jyoti Trivedi, Mohammad Yasir, Rahul K. Maurya and Alok Shiomurti Tripathi. Aptamer-based Theranostics in Oncology: Design Strategies and Limitations. BIO Integration 2024; 5: e993.
DOI: 10.15212/bioi-2024-0002. Available at: https://bio-integration.org/
Download citation
© 2024 The Authors. This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See https://bio-integration.org/copyright-and-permissions/
Abstract
Cancer remains a major global health burden, necessitating innovative approaches for improved diagnosis and treatment. Aptamer-based theranostics have gained attention in the field of cancer research and treatment. Aptamers can be used as targeting ligands for the delivery of therapeutic agents to cancer cells, as well as for the detection and imaging of cancer, due to high binding affinity and selectivity. Aptamers are also being investigated as anticancer drugs. Specifically, aptamers serve as a tool for controlling protein activity via protein-protein and protein-ligand interactions. The aptamer-exosome technology improves aptamer targeting. To gather relevant data, we searched scientific databases, including PubMed/Medline, Google Scholar, Wiley, Web of Science, and Springer. Despite challenges, theranostics, environmental monitoring, biosensing, and other fields could benefit from the use of aptamer technology. This review discusses the standard methods for producing aptamers, including green aptamers, and potential applications in diagnostics. Aptamers are useful in biotherapy and as anticancer drugs, and this article gives a thorough overview of both with examples. We also covered aptamer-exosome technologies, aptasensors, and their diagnostic and therapeutic applications. We investigated recent systematic evolution of ligands by exponential enrichment (SELEX) methodologies with a focus on carrier materials and technical advances, and discuss the difficulties in creating aptamers that are more practical, highly efficient, and stable.
Keywords
Aptamer, exosome technology, SELEX, theranostics.
Introduction
Cancer causes death worldwide with an estimated 10 million new cases of cancer diagnosed every year. Because the prognosis for some types of cancer is still poor, ongoing research into the causes, prevention, and treatment is helping those who are affected [1]. Nanotechnology has emerged as a promising field in the development of novel cancer theranostics by combining diagnostic and therapeutic modalities [2]. Aptamer-based theranostics have gained attention in the field of cancer research and treatment.
Diseases that can penetrate or spread across various body parts because of aberrant cell proliferation are collectively referred to as cancer. There are many types of cancer, the most common forms of which are breast, prostate, bladder, and brain. The possible cancer treatments include surgery, radiation, chemotherapy, immune therapy, and targeted delivery systems. Prevention strategies involve lifestyle changes. Cancer is becoming more widely known and understood despite the difficulties it causes and various advances in technology. Therapies change the outlook of patients more on the positive side and include a greater quality of life for those who are impacted [3]. Treatments strategies, like chemotherapy and radiotherapy, affect not only cancer cells but also healthy cells, leading to various adverse effects, such as fatigue, nausea, hair loss, and immune suppression [4].
Furthermore, the efficacy of these treatments are limited by cancer cell acquired resistance. However, aptamer-based theranostics have the potential to overcome these limitations. By specifically targeting cancer cells, aptamer-based theranostics minimize off-target effects and reduce toxicity to healthy cells. Moreover, aptamers can be engineered to overcome acquired resistance by targeting alternative pathways or biomarkers that are still present in resistant cancer cells. Furthermore, combining aptamer-based theranostics with other treatment modalities, such as chemotherapy or immunotherapy, can enhance the overall efficacy of cancer therapy [5].
Aptamers are short, target-specified, single-stranded oligonucleotides with high selectivity and affinity for target molecules. Aptamers are akin to deoxyribonucleic acid (DNA) or ribonucleic acid (RNA). Aptamers are made in vitro by applying the systematic evolution of ligands by exponential enrichment (SELEX) method. Aptamers can replace or substitute antibodies in some applications due to excellent affinity and selectivity. Aptamers are easily modifiable with functional groups to create novel sensing and targeted delivery platforms with numerous possible uses, including diagnostics, therapy, and biosensing [6].
Aptamers are used as biosensors to detect small molecules and proteins, for targeted drug delivery, in diagnostic assays to identify cancer markers, infectious diseases, and genetic disorders, and as a tool to change the activity of proteins by determining how proteins interact with other proteins and ligands [7].
This introduction provides a brief overview of aptamers and aptamer applications. Further, we discuss perspectives and related challenges in aptamer technology design for cancer and targeting cells in theranostics [8].
Aptamer design
A unique nucleotide sequence that has a binding to the target site is chosen from a library of randomly produced sequences to create aptamers. Various techniques are used in the selection process and the aptamer synthesized is stable under various environmental conditions [9].
Short single-stranded oligonucleotides, which are known as nucleic acid aptamers, have high selectivity and affinities towards binding target sites (low molar-picomolar range) that are comparable to antibodies [10]. Nucleic acid aptamers combine with a variety of targets, including whole cells [7, 11], proteins [7, 11], small molecules [12], metal ions [13], and viruses and bacteria [14, 15]. Aptamers are preferred compared to antibodies owing to their smaller size, faster selection in in vitro, production of chemicals without the use of cells, improved tissue perforation, and lower immunogenicity [7].
Aptamers having high affinity and specificity towards various targets and have great potential as anticancer drugs/agents, so an overview of aptamer roles according to target classification is discussed below.
Proteins: Aptamers can target specific proteins, such as growth factors or cell surface receptors, that are implicated in the progression of cancer. Aptamers can block signalling pathways essential for tumour growth and metastasis by attaching to these proteins.
A flexible approach to treatment is provided by aptamers, which target proteins implicated in the development of cancer. For example, vascular endothelial growth factor (VEGF) is an important angiogenesis mediator that is required for tumour growth and metastasis. Some aptamers, including pegaptanib, have been created to exclusively bind to VEGF, preventing VEGF from interacting with receptors and preventing angiogenesis [16]. Similarly, overexpression of the epidermal growth factor receptor (EGFR) is frequently observed in a variety of malignancies, in which EGFR promotes cell survival and proliferation. Aptamers, such as E07, have demonstrated encouraging outcomes in EGFR signalling inhibition and cancer cell death induction [17]. Aptamer AS1411, for example, targets nucleolin, a protein that is overexpressed in cancer cells, causing apoptosis and inhibiting the growth of cancer cells [18].
Small molecules: Small molecules linked to the onset of cancer or resistance to treatment can be bound by aptamers through engineering. As inhibitors to prevent these molecules from functioning, these aptamers can also serve as drug delivery vehicles.
Innovative therapeutic options are provided by aptamers that target small molecules implicated in the development of cancer or resistance to treatment. For example, adenosine triphosphate (ATP) is frequently dysregulated in cancer cells and is essential for the metabolism of cellular energy. Aptamers that target ATP, like aptamer-ATP-Cy3, have been created to specifically transport drugs to cancer cells and increase the effectiveness of chemotherapy [19]. Furthermore, small molecule medications can be coupled with aptamers that target cell surface receptors, including nucleolin, for targeted distribution that reduces off-target effects and improves therapeutic outcomes [20].
Metal ions: Metal ions have a variety of functions in cancer biology. Aptamers can be tailored to bind to specific ions for theranostic purposes. Aptamer-based sensors (Aptasensor) have been developed for the purpose of detecting metal ions linked to cancer, such as lead or cadmium. Aptamer-based sensors have been created, which potentially provide tools for early diagnosis [21].
Aptamers that target ions have the potential to be used for both diagnosis and treatment. Metal ions have a variety of roles in cancer biology. Aptamer-based sensors that target metal ions linked to cancer, including gold, lead or cadmium, present opportunities for personalised treatment and early cancer diagnosis [22].
Viruses: Aptamers can be engineered to specifically target viral components that are essential for infection or reproduction in viral infections that have been linked to malignancies.
Aptamers that target viral components have special therapeutic prospects. For example, aptamers that target human papillomavirus (HPV) proteins, like E6 or E7, have been created for targeted therapy because HPV infection is a major risk factor for cervical cancer [23]. Furthermore, aptamers that target viral proteins necessary for viral replication, such as the HIV reverse transcriptase, offer promise for antiviral treatment as well as the prevention of malignancies linked to viruses. Aptamers have been created against HIV-1 envelope glycoprotein gp120 and may be used as antiviral medicines to prevent cancers linked to viruses [24].
Bacteria: Some cancers are related to persistent bacterial infections and aptamers can be used to target bacterial components or toxins connected to carcinogenesis.
Aptamers that target bacterial components or toxins present new therapeutic options. For example, Helicobacter pylori infection has been linked to gastric cancer and aptamers that target toxins or bacterial surface proteins, like VacA or CagA, have been created for targeted therapy [25]. Furthermore, by reducing chronic inflammation linked to bacterial infection, aptamers that target bacterial toxins, such lipopolysaccharide (LPS), have the potential to prevent cancer [26].
The methodology for purifying and describing the aptamer should be incorporated into the aptamer design, as discussed below.
Traditional
SELEX methodology
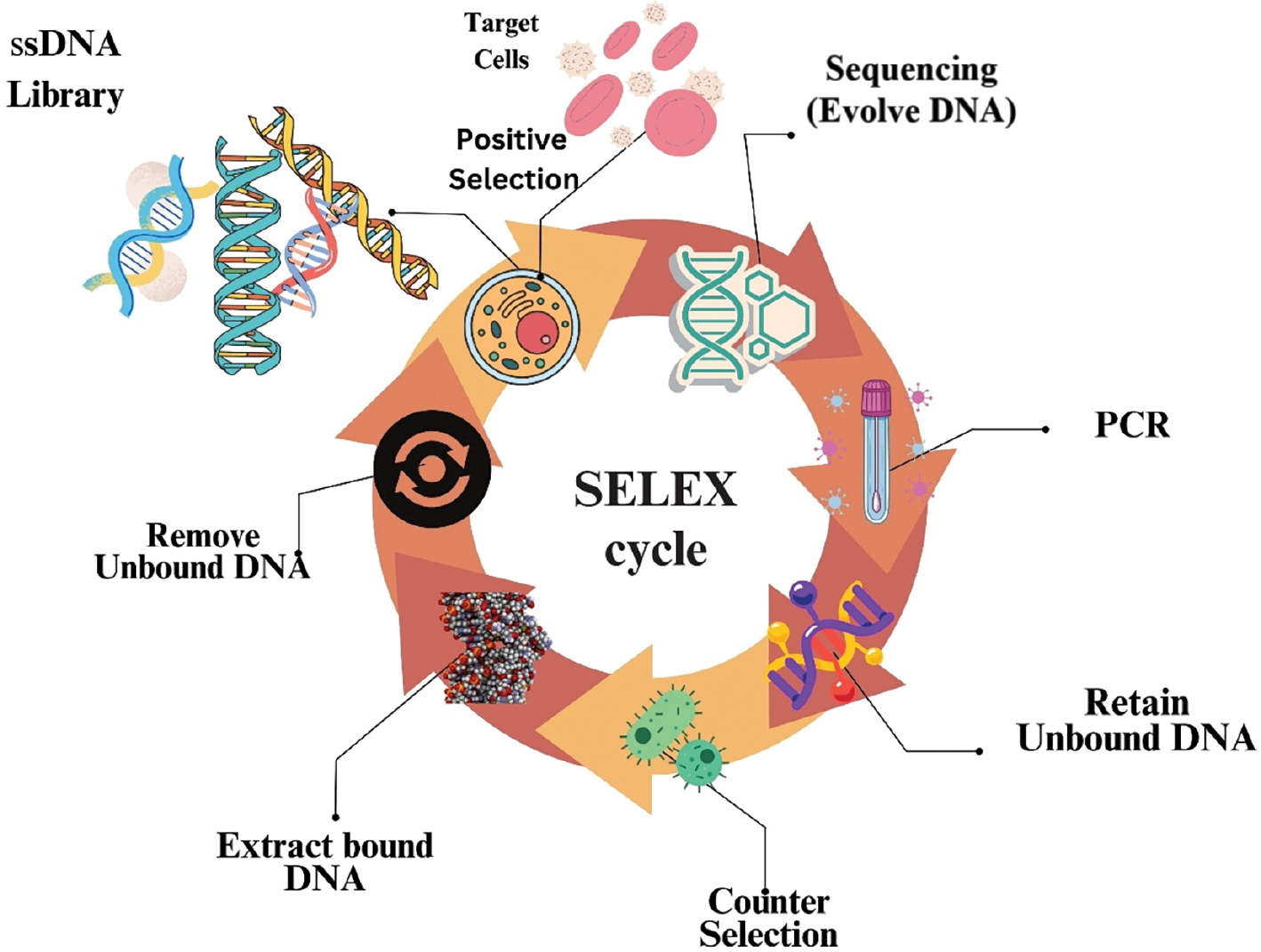
Three distinct teams used a selection technique (SELEX) in the 1990s to isolate RNA aptamers [11, 27]. The four general selection steps are typically involved in technology, as shown in Figure 1.
Figure 1 Flowchart representing the various steps of SELEX technology.
- The target interest and a baseline library of DNA and RNA are combined, which marks the beginning of the selection cycle. A library typically contains up to 1015 chosen 20–60 nucleotide sequences that are flanked at the 5′ and 3′ ends by fixing primer regions.
- Target- and un-bound sequences are separated after incubation using a variety of partition methods.
- For the next selection cycle, a new library is created using the bound sequences that have been recovered and reamplified.
- Prior to utilizing polymerase chain reaction (PCR) procedures, such as transcription and amplification to form a new RNA library, retrieved RNA patterns need to be converted into cDNA using the reverse transcription technique, whereas new DNA libraries can be generated directly using the PCR technique [7].
After 2–15 iterations of the selection cycle, analysis of the RNA patterns utilized to recognise specific patterns from the collection is enhanced. Throughout the selection procedure, modifications are made in the library with respect to incubation time, target ratio, and buffer composition. Selection stringency improves the enhanced target-bound sequences. Various SELEX technologies targeted at select molecules are summarised in Table 1.
Table 1 Various SELEX Technology Targeted to Selected Molecules with Examples
| SELEX Technology | Selected Molecule | Examples | References |
|---|---|---|---|
| DNA-SELEX | DNA-binding proteins, DNA aptamers | HMGB1 protein, TATA-binding protein, DNA aptamers for cocaine | [11, 131] |
| RNA-SELEX | RNA-binding proteins, RNA aptamers | HIV-1 Rev protein, nucleolin protein, RNA aptamers for ATP | [10, 14] |
| Chem-SELEX | Small molecules, peptides, proteins | ATP, streptavidin, tau protein | [132, 133] |
| Cell-SELEX | Whole cells, cell surface proteins | Leukaemia cells, breast cancer cells, T lymphocytes | [134, 135] |
| Protein-SELEX | Proteins, peptides | TGF-β1 protein, PDGF-BB protein, peptides that bind to fibrinogen | [136, 137] |
| Spiegelmer-SELEX | Mirror-image oligonucleotides, mirror-image aptamers | Thrombin protein, VEGF protein, mirror-image RNA aptamers for IFN-γ | [10, 138] |
Green aptamer
Cell imaging studies are crucial to understanding cellular physiology. Green fluorescent protein (GFP) and targeted cells (proteins) have long been studied for their interactions, positioning, claims, signals, and functions, which are key parts of cellular imaging applications. Yet, a large amount of RNA research has begun to broaden the function of RNAs, leading to the creation of cutting-edge intracellular imaging technologies, including fluorogenic aptamers, such as those found in spinach [28], broccoli [29], mangoes [30], and corn [31], which cause fluorescence to be activated when target fluorophores are bound. These green or fluorescent aptamers can be made into imaging tags to determine how RNA moves inside cells or as biosensors to track metabolites inside cells in a laboratory setting.
Spinach as a fluorogenic
Green fluorescence is caused when the small molecule, fluorophore 3,5-difluoro-4-hydroxy-benzylidene-imidazolinone (DFHBI) and its variations, including DFHBI 1T and DFHBI 2T, connect to the spinach fluorogenic RNA aptamer [28]. The spinach-DFHBI complex was determined using crystal structure analysis and this information was used to investigate the exact mechanism underlying fluorescence activation.
The spinach-DFHBI complex co-crystal structures were shown to have an unexpected quadruplex core coupled with a combination of a mixed base and two G-tetrads. The unpaired guanine residue (G31) is encased by a base-triple-inserted DFHBI. In the fluorophore-free state, in contrast, the base tier of the quadruplex is rearranged with the fluorophore collapsing binding site, in which the helical stack can grow to G31 [32].
Broccoli as a fluorogenic
The broccoli isolation technique was created to increase fluorogenic aptamer functionality and specificity. Following an initial round of traditional aptamer selection, fluorescent RNA aptamers were introduced into plasmids using experimental techniques. Fluorescence-activated cell sorting was utilized for isolation of fluorescent cells after cloned plasmids were transformed into E. coli for DFHBI treatment. The green aptamer, broccoli, which is fluorogenic, was successfully obtained using this method and displayed vigorous folding in low concentrations of salt, enhanced thermal stability, and higher fluorescence than spinach [29]. Broccoli was dimerized into tdBroccoli after being shortened to tBroccoli for any further in vitro imaging. The dimerized tdBroccoli fluoresced with twice the intensity of broccoli [29]. Without a tRNA scaffold, broccoli or dBroccoli display greater fluorescence intensity than a tRNA scaffold. In transfected HEK293T cells, microscopy was used to compare fluorescence activation [29].
Mango as fluorogenic
The mango RNA aptamer exhibits orange-coloured fluorescence in significant levels. Derivatives of the fluorophore thiazole orange (TO1) increase fluorescence by a factor of 1100. In an A-form duplex, two-tiered all-parallel G-quadruplexes were anticipated to fold the structure of mango. Three antiparallel quinines creating G-quadruples were revealed by the crystal structure investigation of the mango-TO1 complex [33]. The G-flat quadruplex face on which TO1 is bound, stabilizes the mango structure and turns on the orange fluorescence.
For imaging purposes, equimolar quantities of RNA mango were used to directly inject TO1 into the syncytial gonads of Caenorhabditis elegans (C. elegans). The gonad nuclei were shown to have a high fluorescence composition [30].
Corn as a fluorogenic
Yellow fluorescence is triggered by the corn RNA aptamer, which combines to form 3,5-difluoro-4-hydroxynenzylideneimidazolinone-2-oxime [i.e., DFHO] [31]. Corn exhibits much greater photostability than broccoli. Fluorescence is visible at 320 ms imaging periods but broccoli is only visible at 160 ms [31]. In contrast to the broccoli-DFHBI complex, which loses >50% of fluorescence past 200 ms, the corn-DFHO combination in the irradiation trials reveal very little fluorescence loss after 10 s. Corn is therefore regarded as a newly emerging aptamer that is fluorogenic and acts as a biosensor. The corn aptamer yields three primary subtypes (tRNA, 5S RNA, and U6 [Pol III promoters]). The tRNALys scaffold plasmid facilitates the assessment of viability as a fluorescent RNA tag that is genetically encoded [34].
Aptamer-exosome technology
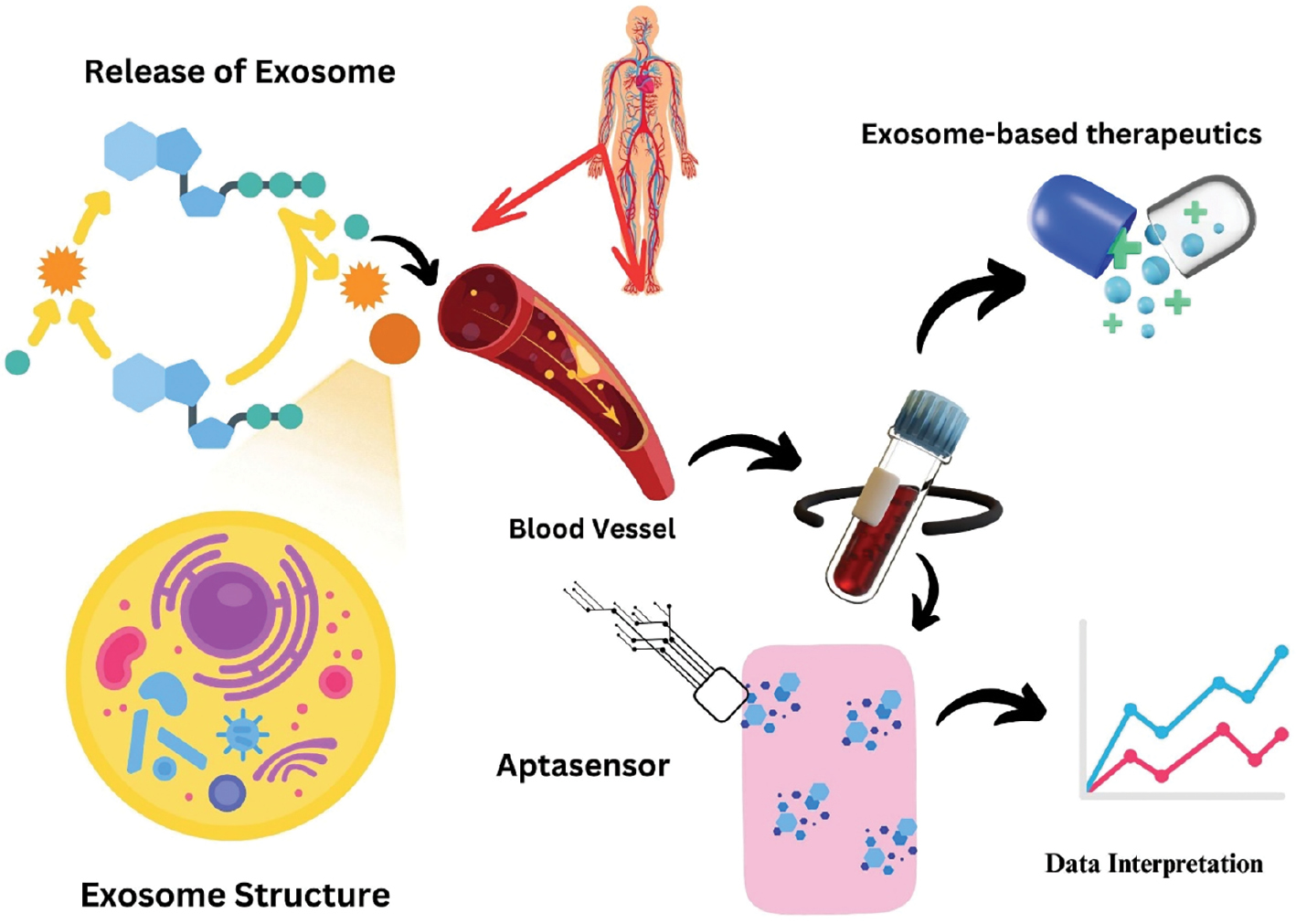
All live cells secrete extracellular vehicles (EVs), which are membrane-bound bionanoparticles [35]. Based on the mechanism underlying synthesis and size, EVs are categorised as exosomes, oncosomes, exomers, apoptotic bodies, or microvesicles [36]. EVs have progressed from the early days as “platelet dust” [37] to becoming an important intermediary for in vivo interaction across some cellular entities, tissues, and cross-kingdom molecules [38]. Exosomes and microvesicles have been identified after long-lasting cell biology and biochemistry research. Exosomes have particle sizes ranging from 50–150 nm with surface biomarkers known as tetraspanins [CD63, CD81, and CD9] [39], while microvesicles have particle sizes ranging from 100–1000 nm and annexin A1 as a distinctive biomarker.
Exosomes carrying cargo acquired from the cells of origin have been discovered in blood and different body fluids, circulating in a variety of cell types. These developments have created the groundwork for exosomes to become a cutting-edge source for identifying new biomarkers. Tumour-susceptibility gene 101 (Tsg101), heat shock proteins, the endosomal sorting complex needed for transportation (ESCRT-3) binding protein, Alix, and the major histocompatibility complex, both class I and class II composites, are known to be carried by exosomes [40].
Exosome attachment and fusion with the plasma membrane of the recipient cell may be facilitated by the additional attachment of molecules to the surface, such as lymphocyte function-associated antigen-1 (LFA-1) integrin and intercellular attachment of molecule-1 [ICAM-1 {often called CD54}] [41]. In addition, the fact that exosomes contain a lot of transmembrane proteins, such as EGFRs and epithelial cell adhesion molecule (EpCAM), suggests that exosomes arise from living structures. These proteins are used as excellent biomarkers because the proteins are connected to the typical pathophysiology and pathogenesis of numerous diseases [42].
Exosomes are extremely durable in a variety of in vivo and in vitro settings because the membranes are significantly abundant in lipid rafts [43]. These proteins are used as excellent biomarkers because they are connected to the typical pathophysiology and pathogenesis of numerous diseases [44], as shown in Figure 2.
Figure 2 Extracellular vesicles as theranostics.
Capturization-aptamers to exosomes
Exosomes include information about the prognosis and development of a specific condition in addition to the parent cell, making exosomes useful diagnostic indicators. For example, exosomes produced by cancer cells may contain membrane proteins vital to the development of the disease.
Membrane proteins are involved in the development of cancer. Programmed death ligand 1 (PD-L1) has been shown to suppress CD8+ T lymphocytes and promote tumour growth, based on a recent study involving exosomal PD-L1 derived from metastatic melanoma cells [45]. Integrins, such as α6, αv, and β1, that are present in exosomes produced by cancerous cells can be used to forecast the tumour stage and differentiate between different types of cancer, including the breasts, kidneys, colon, and ovary [46] because more of these proteins were released by aggressive progenitor cancerous cells.
Research has demonstrated that exosomes can pass through the blood-brain barrier, at least in some cases. Therefore, the quantity and composition of exosomes in plasma and cerebral fluids can reveal important details about the pathogenesis and development of neurodegenerative disorders [47]. The biological properties of exosomes enable vesicles to efficiently transport therapeutic compounds. This homing effect of exosomes suggests that therapies using exosomes may have better results in preventing cancer in the cancer cells from which exosomes arise [48].
Technology in theranostics
Exosome diagnostics using aptamers
Aptamers can be chosen to precisely bind with biomarkers on exosome surfaces and confine the exosomes to enhance the enrichment efficiency of the exosome and facilitate exosome isolation. The following sections provide an overview of the development of aptamer-guided exosome diagnostic tools.
Exosomes can be easily captured using peptide/RNA/DNA aptamers
A diagnostic tool, the aptamer-guided exosome-capturing nanoplatform technique, is used by choosing an aptamer contrary to an overexpressed polypeptide on exosomes. An aptamer that connects to the extracellular domain of HSP70 was developed by Garrido et al. [49] with the aim of targeting the A8 peptide, which is increased in numerous cancers. HSP70-positive exosomes were discovered using this aptamer in urine samples from patients with breast, lung, and ovarian cancer [49].
Exosome levels in the blood and urine can be measured. After multiple iterations of SELEX, Murakami et al. [50] chose two RNA aptamers with lengths of 55 and 30 nucleotides (nt) each. According to surface plasmon resonance (SPR) analysis, the 55- and 30-nt aptamers have similar levels of attraction for exosomes. Circular dichroism spectroscopy showed that the two aptamers are capable of forming G-quadruplex structures in between loop regions that are supported by K+ or potassium ions [50]. Sun et al. [51] devised ways to use DNA aptamers and DNA to separate exosomes of a specific size and identify the surface proteins at the same time in different studies. Stage II breast cancer patients with varying HER2 expression patterns and different breast cell lines could be distinguished using a machine learning algorithm implemented to assess exosomal size and marker signatures [51].
Aptasensor
Aptasensors have developed into a crucial diagnostic tool for identifying cancer-derived exosomes due to high sensitivity, quick response, accessibility, and low volume of sample [51].
Other diagnostic methods based on labs-on-a-chip technology
The rationale behind aptamer-guided exosome diagnostic tools is that aptamer-guided exosome diagnostic tools selectively attach to exosomes that carry disease-related information. Aptamers are mixed with polymers and other inorganic substances to make a platform that is built right into lab-on-a-chip exosomal diagnostic tools. The ExoAPP test, an aptamer nanoprobe-based exosome profiling technique, was used to characterise the surface proteins present in exosomes and detect cancer-derived exosomes [52].
Exosome therapeutics using aptamers
Exosomes with paclitaxel, an anticancer medication frequently used in chemotherapy regimens, were described by Wan et al. [53] as a novel platform for clinical applications in treating cancer.
The cholesterol-PEG-covalently-conjugated nucleolin targeting aptamer AS1411 was then fixed to murine dendritic cell membranes. Then, to produce aptamer-guided exosomes, these cells were physically extruded. This method used extrusion to make 3×1010 aptamer-guided exosomes with a diameter of 105 nm from approximately 1×107 cells/h. Because of amphiphilicity and corresponding stiffness, cholesterol-PEG2000 was chosen because cholesterol-PEG2000 stabilizes exosomes at the lipid bilayer membrane through hydrophobic contacts. The technique used to create cholesterol-PEG2000-aptamers could be potentially applied to the mass manufacturing of specific exosomes released through immune cells for the treatment of various malignancies. The cells needed to generate exosomes are collected from the patients who require therapy, making this procedure relatively more secure than cell-based immunotherapy, such as CAR T-cell therapy.
Such derived exosomes can be used to be genetically engineered without drug loading [53]. An alternate strategy involving reprogramming of exosomes leads to effective targeted exosome delivery and tumour suppression, which can be achieved by controlling surface ligand presentation and siRNA/miRNA loading on exosomes with an RNA aptamer [54].
Aptasensor
Having high affinity, stability, ease of modifying kinetic parameters, relatively quick creation without the use of animals, and a wide range of targets from tiny molecules to entire cells, aptasensors are essentially antibody-based sensors [55]. To identify cancer, there are numerous optical, electrochemical, and colorimetric aptasensor techniques. This section will concentrate on current developments in aptasensors for the sensing and identification of cancer, with a focus on developments.
Electrochemically activated aptasensors
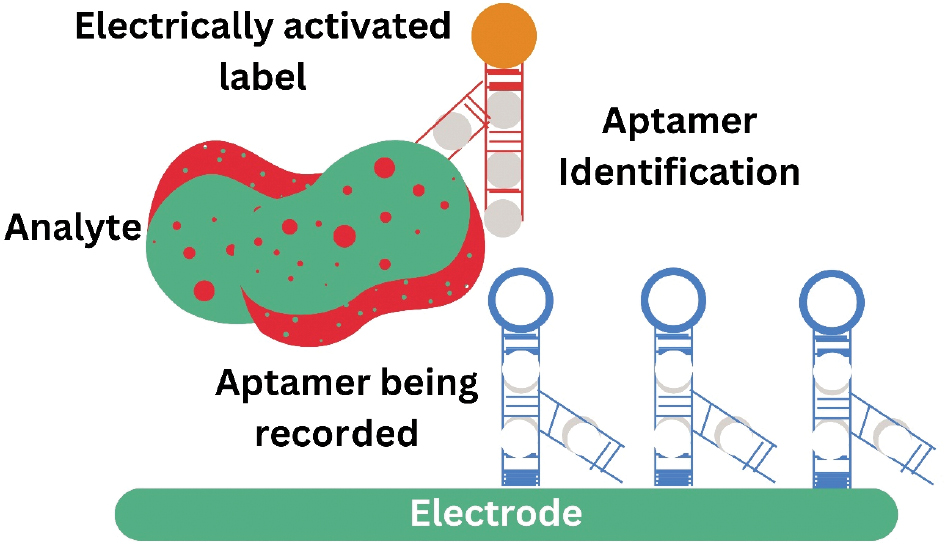
Sandwich-style aptasensor
The sandwich-style aptamer system basically consists of an electrode base and two aptamers (Figure 3). The analyte is captured and immobilised by a capturing aptamer coupled to an electrode surface, whereas another aptamer named secondary aptamer identifies and binds different parts of the analyte base and forms an aptamer-analyte-aptamer sandwich. The electrode [56] can be used to detect an electroactive label present in the secondary aptamer, such as cadmium sulphide quantum dots [57], glucose dehydrogenase [58], or gold nanoparticles [AuNPs] [59]. Numerous sandwich-based detection systems were designed due to the relative simplicity in cancer cell targeting.
Figure 3 Representation of sandwich-style aptasensors activated electrochemically.
Zhang et al. created an aptasensor that is electrochemically activated by means of an aptamer against the surface glycoprotein, Mucin1 (MUC1), which is markedly overexpressed in many cancers. A similar MUC1 aptamer-conjugated magnetic bead was caught by an extra lectin-based nanoprobe functionalized on AuNPs, which then attracted cells that express MUC1 [60]. In this experiment the reduction of silver ions by gold caused voltage fluctuations. Utilizing electrochemical stripping analysis, these voltage differences were read to determine the amount of MUC1 expression, and potentially, a carcinogenic diagnosis.
Label-free aptasensors
The properties of aptamers, such as the distinctive attributes changing upon binding to a target, enhance resistance triggered by the creation of double-stranded DNA and decrease signalling when an electrode electroactive group is moved by aptamer binding, have been harnessed to create label-free, electrochemically activated aptasensors [56, 61]. Various electrochemically activated aptasensors, which are label-free, have been created against cancerous cell targets and aptasensors that benefit from the conformational shift that occurs as aptamers attach to their targets.
Wang et al. [62] recognized MCF-7 breast malignant cells using a polyadenine-modified aptamer system using the voltage drop caused by target binding that is detected by differential pulse voltammetry.
Aptasensor-activated fluorescence
The first aptamer-based biosensor was created using a fluorescent aptamer against human neutrophilelastase (HNE) in 1996. It was shown that the HNE fluorescent aptamer is equally efficient as an antibody in detecting HNE on beads. However, the HNE fluorescent aptamer also had additional benefits, such as the capacity to include functional groups faster, a smaller size that makes internalisation easier, the ability to identify intracellular targets, less off-target binding, and higher storage stability [63].
Herr et al. [64] created a fluorescent sandwich system for the identification of cancerous cells using aptamers coupled with fluorescent nanoparticles. Herr et al. [64] created a useful technique for therapeutic usage using magnetic nanoparticles to assist with taking these cells out of entire blood samples.
Chen et al. [65] developed a multiplexed detection technique in 2009 that uses aptamer-conjugated Forster resonance energy transfer (FRET) using silica nanoparticles to identify several cancer cell targets. Numerous fluorescent aptasensors have been created to detect cancer. Fluorescent aptasensors are used to detect cancer indicators in addition to malignant cells. VEGF is one of the markers that a fluorescent aptamer was initially intended to detect.
Freeman et al. [66] reviewed several optical aptasensor techniques that exploit the structural change of an anti-VEGF aptamer when an anti-VEGF aptamer interacts with its target. Specifically, some sensing approaches were evaluated based on fluorescence resonance energy transfer, luminescence emission via chemical means, and luminescence resonance energy transfer methodologies that enable target detection [66].
Cho et al. [67] used the optical phenomenon known as nanoplasmonic sensing, in which fluorophore intensity is altered when the fluorophore encounters free electrons on a metal surface to create a one-step method for VEGF165 detection. Using this technique, cyanine (Cy3) was used to mark the surface of AuNPs with anti-VEGF aptamers. The aptamer conformation is modified when the aptamer is bound to VEGF, releasing the aptamer from the AuNPs and causing a substantial drop in fluorescence intensity [67].
Aptasensor-activated colorimetry
Several aptamer-based colorimetric assays facilitate the identification of cancer markers, allowing for easy and quick recognition of targets that range from tiny metal ions to complex structures, such as proteins.
Xu et al. [68] designed a colorimetric biosensing technology designed to identify the proto-oncogene (K-Ras) using a DNA molecular machine. The main part of the device is a hairpin detector that can find K-Ras and connect to a primer-linked polymerization unit to make an anti-hemin aptamer. This anti-hemin aptamer facilitates the conversion of the colourless substrate, 2,20-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) [ABTS], into a green-coloured form, allowing visual identification without any equipment. This process activates a DNAzyme that mimics the activity of horseradish peroxidase [68].
Ahirwar and Nahar et al. [69] designed a nanoparticle-based colorimetric aptasensor system for the recognition of the human oestrogen receptor alpha (ERα), a usual marker in breast cancer.
Theranostics benefits of aptamer
Aptamers serve a purpose in theranostics because of a dual function (therapeutic and diagnostic purposes). Aptamers provide high affinity and specific selectivity for target molecules that have the capacity to bind to certain sites, which allows aptamers to be used as delivery systems for medicinal drugs, allowing for tailored or conjugation therapy, as well as for diagnostic purposes by identifying biomarkers. By optimizing therapeutic outcomes and offering real-time monitoring of treatment response, this dual functionality advances the theranostics use of medicine [70].
Chemotherapy
One of the limitations of traditional cancer treatments, such as chemotherapy and radiotherapy, is the potential for significant side effects. These treatments often affect not only cancer cells but also healthy cells, leading to various adverse reactions, such as fatigue, nausea, hair loss, and immune suppression. Furthermore, the efficacy of these treatments can be limited by cancer cells acquired resistance However, aptamer-based theranostics have the potential to overcome these limitations. By specifically targeting cancer cells, aptamer-based theranostics minimize off-target effects and reduce toxicity to healthy cells. Moreover, aptamers can be engineered to overcome acquired resistance by targeting alternative pathways or biomarkers that are still present in resistant cancer cells [71].
Various drugs that have been used in chemotherapy studies are below.
Doxorubicin (DOX)
DOX is often used in several cancer treatments. When small molecules are coupled to DOX, the resistance caused by cancer cells is decreased, which therefore enhances the efficiency of chemotherapy [72]. As a constituent of the anthracycline family, DOX has the Food and Drug Administration (FDA) approval. DOX inhibits DNA replication by intercalating DNA and subsequently causes adverse reactions, such as irreversible cardiomyopathy brought on by oxidative stress, myocardial fibre loss, apoptosis, and downregulation of genes that regulate contractile proteins [1].
The AS1411 aptamer is a G-quadruplex-structured antiproliferative DNA aptamer that was created by targeting nucleolin, a proteinaceous structure, which is abundant in cancerous cells. This oligo-nucleotide, the first aptamer, provided the basis for clinical trials involving a variety of cancer treatments and was subsequently used in additional studies to improve the effectiveness of the treatment against cancerous cells [73]. Details of the clinical trials based on aptamer along with its NCT number are shown in Table 2.
Table 2 Clinical Investigation Done on Aptamer in the Field of Oncology
| S. No. | NCT Numbera | Drug/Intervention | Cancer | Study Type | Volunteers/Clinical Trial | Summary | Status | References |
|---|---|---|---|---|---|---|---|---|
| 1. | NCT01830244 | Nab-paclitaxel (Abraxane) | Breast | Interventional Primary purpose: Treatment |
60 participants Phase 2 |
This project aims to assess tailored primary systemic therapy using sequential nab-paclitaxel, epirubicin, and cyclophosphamide for early breast cancer. It is an open-label phase II clinical trial hypothesizing that this regimen is feasible and yields high response rates. The study planned to enroll 60 patients, including 40 with chemotherapy-sensitive tumours and 20 with ER-positive tumours, potentially responsive to hormonal treatment. The target group was women with early breast cancer eligible for primary systemic therapy. The primary outcome was overall response rate in the breast. Secondary outcomes included response rates in axillary lymph nodes, safety, tolerability, and breast conservation rate. Participants underwent genotype testing to explore chemotherapy sensitivity prediction, with results not affecting therapy selection. Optionally, patients can contribute tumour tissues for laboratory studies on cancer-initiating cells and aptamer-based tumour targeting. | Unknown | [139, 140] |
| 2. | NCT00056199 | EYE001 | von Hippel-Lindau disease | Interventional Primary purpose: Treatment |
5 participants Phase 1 |
von Hippel-Lindau (VHL) disease is an inherited disorder causing various benign and malignant tumours across multiple organs. Retinal angioma, a common symptom, can lead to vision loss due to retinal edema. Traditional treatments, such as photocoagulation or cryotherapy, may not work for all lesions. The study aimed to test the effectiveness of anti-VEGF therapy (EYE001) on retinal angiomas associated with VHL in five patients. The patients received 6 injections over 30 weeks and were monitored for improvements in vision, retinal health, and any adverse effects. | Completed | [141] |
| 3. | NCT00740441 | AS1411 | Metastatic renal cell carcinoma | Interventional Primary purpose: Treatment |
30 participants Phase 2 |
The aim of this studywas to evaluate the efficacy of AS1411 in patients with metastatic renal cell carcinoma. | Unknown | [142] |
| 4. | NCT01486797 | NOX-A12 | Chronic lymphocytic leukaemia | Interventional Primary purpose: Treatment |
28 participants Phase 2 |
The purpose of this study was to evaluate the safety and efficacy of NOX A12 in combination with a background therapy of bendamustine and rituximab chemotherapy in previously treated patients with chronic lymphocytic leukaemia. | Completed | [143, 144] |
| 5. | NCT03385148 | 68Ga-Sgc8 | Colorectal | Interventional Primary purpose: Diagnostic |
70 participants Early Phase 1 |
The protein, tyrosine kinase-7 (PTK7), is overexpressed in various types of human cancers. As a specific imaging agent of PTK7, 68Ga-Sgc8 was investigated in this study to assess its safety, biodistribution, and dosimetric properties in healthy volunteers, and to preliminarily evaluate its application in colorectal patients. | Unknown | [145] |
| 6. | NCT06258330 | AM003 | Solid tumour | Interventional Primary purpose: Treatment |
15 participants Phase 1 |
This is a phase 1, first-in-human study to assess the safety and tolerability of AM003 in patients with locally advanced and metastatic solid tumours. | Recruiting | [146] |
aNCT Number- ClinicalTrials.gov Identifier
Li et al. [74] worked on an advanced aptamer system (AS1411) that delivers DOX to patient malignant breast cells (MCF-7). The hearts of mice bearing the MCF7 tumour showed no indication of drug accumulation. Micelle technology significantly increased AS1411 affinity for MCF-7 receptors. The reduced accumulation of DOX in cardiac cells led to a reduction in cardiac tissue necrosis and damage, which provided a feasible avenue for aptamer-based therapy. Moreover, AS1411 exhibited anticancer activity and a decreased level of cardiotoxicity by DOX in MCF-7/ADR tumour-having mice, as well as decreased resistance to multiple drugs mediated by glycoprotein-P (P-gP) discharge [74], as shown in Table 3.
Table 3 Various Drugs with Aptamers Specifications with Cell Line used in Chemotherapy Studies
| S. No. | Name of Drug | Aptamer | Cell Line | Cancer Type | References |
|---|---|---|---|---|---|
| 1 | Doxorubicin (DOX) | AS1411 | MCF-7 | Breast | [35, 94, 96] |
| MUC-1 | LNCaP | Prostate | |||
| SL2B | HT-29 | Human colorectal adenocarcinoma | |||
| 2 | Docetaxel (DTX) | AS1411 | C6 Glioma | Brain | [37] |
| 3 | Epirubicin (Epi) | Anti-MUC-1 | CHO | Ovarian | [36, 37] |
| MCF-7 | Breast | ||||
| 4 | Gemcitabine (Gem) | RNA | EGFR | Pancreatic | [1] |
The same investigation used a bubble generating agent. When the microenvironment is warmed to 42°C, intracellular DOX content rises and potentially harmful drug release occurs, leading to permeability defects in the bilipid layers. The use of novel aptamers that bind and block the proteins necessary for the formation of P-gP is intended to remove restrictions on the therapeutic application of DOX. This maneuver will increase DOX release and decrease carcinoma cell survival without having a deleterious effect on cardiac muscle failure. Trinh et al. [75] also developed AS1411 and DOX into a human model of hepatocellular carcinoma. No negative effects on the heart or endocrine system, as well as no weight loss were noted [75].
Atabi et al. reported that MUC-1 aptamers make DOX persist longer in human prostate cancer cell lines (LNCaP cells). Jing et al. concluded that the double aptamer system decreases cancerous growth in prostate cancer (LNCaP and PC3 cells). The combined effects of DOX and MUC-1 aptamers on MCF-7 malignant cells found in breast tissues were investigated by Jeong et al. Similarly, Sun et al. worked on a human colorectal adenocarcinoma cell line (HT-29) to observe the effect of the SL2B aptamer. These investigations provided a path for the development of aptamers, which resulted in chemotherapy that is administered at lower doses, reducing the afore-mentioned negative effects [76].
Docetaxel (DTX)
DTX is a taxane-family anticancer medication that promotes cell death by depolymerizing tubulin and causing microtubule aggregation. Intravenous DTX has been associated with toxicity in malignant brain tumours [77].
Gao et al. [78] utilized AS1411 as a booster for the DTX anti-glioma impact in the C6 glioma line. Treatment for gliomas is particularly challenging because the blood-brain barrier (BBB) and brain-glioma barrier inhibit the uptake of drugs into tumour cells. Double aptamer systems can be used to improve such investigations, both for BBB detection as well as cancer identification.
Paclitaxel (PTX)
PTX is a chemotherapy agent that treats malignant glioma cells, although reduced solubility in water reduces PTX efficacy. In the interest of promoting enhanced permeability and retention within tumours, aptamers may be used to increase the solubility of PTX. Increasing tumour penetration and retention of macromolecular medications can lessen toxicity to healthy cells [79].
Epirubicin (Epi)
Taghavi et al. [80] used the anti-MUC-1 aptamer, 5TR1, and the augmented Epi medication delivered to CHO cells, a hamster ovary cell line, and MCF-7 breast cancer cells. Epi is a medication from the anthracycline family that when used in high quantities can lead to cardiotoxicity. Novel aptamers may improve MUC-1 glycoprotein recognition to reduce myocardial cell cytotoxicity, improve Epi release to boost DNA intercalation, block topoisomerase II, and increase degradation of cancer cells.
Gemcitabine (Gem)
The RNA aptamer utilized by Ray et al. to increase the efficacy of the chemotherapeutic medication, Gem, targets EGFR-overexpressed pancreatic carcinoma cells. Gem is a nucleoside molecule that has been linked to neutropenia-related illnesses in high concentrations because of normal cell toxicity. Aptamers that affect Gem phosphorylation status during internalisation can be exploited to circumvent the toxicity effect limits. A nucleoside, Gem, that phosphorylates intracellular cells to limit the function of ribonucleotide reductase stops growing cancer cells from lengthening DNA, as shown in Table 3.
These studies demonstrated that unexplored territories remain in aptamer-based chemotherapy. Aptamers can undoubtedly enhance cancer treatment by increasing therapeutic efficacy and/or decreasing adverse effects [1].
Biotherapy
Immuno-theranostic
Melanoma, and non-small cell lung, head, and neck cancer are just a few of the many cancer types that can be treated with immunotherapy, which is a successful and innovative treatment approach. Several biotherapy tools have been developed by integrating several aptamer features with other additional related molecules, such as proteins or reporter groups.
Zhao et al. [81] configured a bivalent RNA aptamer that prevents heat shock factor (HSF1) from interacting with associated DNA promoter regions, down-regulating expression of HSF1 and making carcinomas more susceptible to anti-cancerous drugs. By using a CD28 2′-fluoro RNA aptamer, Lozano et al. created an aptamer in place of Foxp3 [82]. The initial findings indicate that the P60 Foxp3 inhibitor peptide couples alongside a CD28-targeted aptamer to enable peptide delivery to CD28-articulating cells, thereby suppressing regulatory T-cell immune responses when the cells become trapped within the tumour microenvironment.
While formulating combination medicines, aptamers may also be crucial because aptamers may aid in locating the biomarkers responsible for unfavourable reactions. For example, the mesothelial surfaces that are present on lung pleura are home to drug-resistant malignant pleural mesothelioma (MPM), the biomarkers of which lack sensitivity and specificity [83]. The success rate of treatment could be considerably increased by the creation of precise biomarkers for MPM. One of the frequently used MPM markers is CD44, although there is no reliable measurement method for CD44. The creation of aptamers against this marker may aid in the earlier diagnosis of this condition, enabling a more structured course of therapy.
Various organizations are currently focusing on a category of biomarkers, specifically metabolic biomarkers. This category of markers is consistent amongst the four functional levels (genome, metabolome, proteome, and transcriptome) linked to cell function. This approach has been used in colorectal cancer (CRC) therapies. Accumulation of arginine, proline, glutamine, threonine, lactate, phenylalanine, and a few other amino acids have been identified as an important CRC biomarker [84–86]. The abovementioned metabolic biomarkers, in particular those that are released by cells, can all be detected using aptamers. Because the behaviour of tumour cells can be understood with the help of such metabolites and how the tumour cells react, if at all, to traditional remedies, implementing aptamers for quantification of these metabolites can be beneficial in cancer progression studies.
Drug delivery system: aptamers in anticancer
Drug delivery methods have been in use for some time and have indeed advanced medicinal modalities. Nanocarriers are colloidal nano-scale systems that can carry anticancer substances. Drugs and large molecules with small molecular weights are examples of nanocarriers that facilitate therapy by preventing pharmaceutical deterioration, lowering renal clearance, extending half-life, and helping to control kinetic release. Drugs and large molecules with small molecular weights assist with solubility [87]. Among tumours that grow quickly, cancerous cells are usually situated near an endothelium barrier. As a result, nanocarriers containing targeting molecules will adhere to the primary receptors encountered rather than trying to penetrate the entire tumour [88]. Cell-SELEX is best-suited in this scenario. The procedure selects aptamers that might be beneficial for the nanocarriers by attaching nanocarriers to cellular receptors that are stated above and blocking access to the remaining tumour through receptor saturation.
Targeted drug delivery: aptamer-drug conjugate (ApDC)
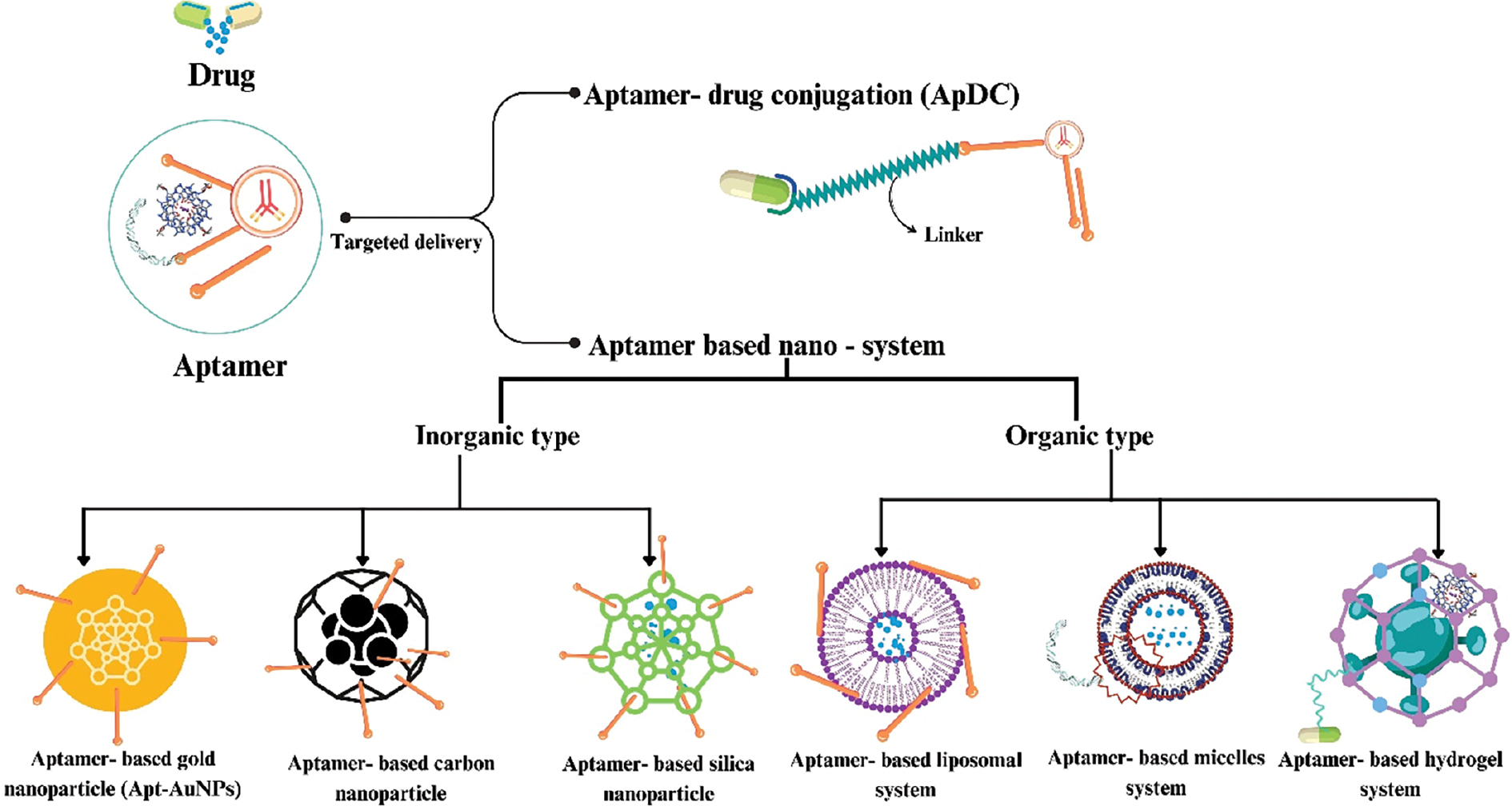
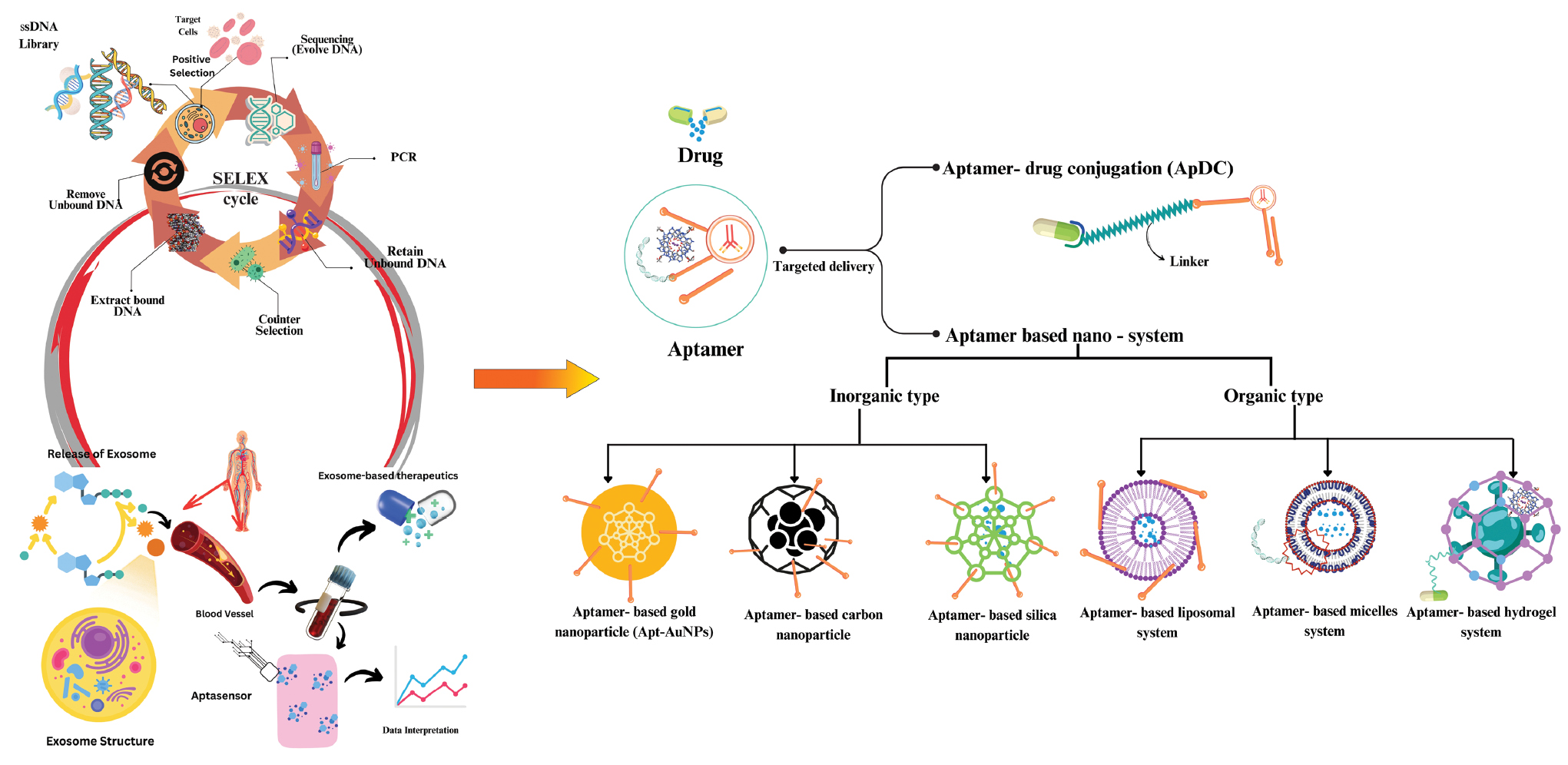
ApDCs have drawn interest as therapeutic agents as well as vehicles for delivering a variety of therapeutic agents, including toxins, small interfering RNAs (siRNAs), microRNAs (miRNAs), and chemotherapeutics [Figure 4] [89]. Conventional ApDCs comprise a combination of aptamers and linkers which are linked via various cleavable or non-cleavable linkers. Compared to antibody-drug conjugates, which have been applied in clinical cancer treatment, ApDCs have several benefits, including quick tissue penetration, facile chemical modification, smaller size, and a cost-effective manufacturing procedure. ApDCs have been produced for a wide range of therapeutic techniques, including immunotherapy and chemotherapy, by targeting biomarkers linked with specific diseases [90].
Figure 4 Schematics of drug delivery methods for aptamers explain aptamer-drug conjugation and aptamer-based nanosystems.
Tan et al. [91] created and synthesized a sgc8-DOX conjugate to deliver DOX in a targeted manner. In these ApDCs, the anticancer drug, DOX, and the aptamer, sgc8, were conjugated at an equal ratio using an acid-labile hydrazone linker, which enabled DOX to be selectively delivered to acidic tumour environments [91].
Aptamers can be used to overcome these difficulties. Receptor-mediated endocytosis, for example, could account for concentration issues. This strategy could be implemented using aptamers that target cell-surface receptors associated with cellular uptake.
Aptamers may accumulate in tumour cells, possessing a major disadvantage to the system, the use of which could potentially be advantageous for drug delivery systems based on lipids. An aptamer against mTOR (Raptor) that performs in a similar manner was chosen by Berezhnoy et al. (4-1BB aptamer-siRNA chimera). When mechanistic target of rapamycin (mTOR) is expressed, mTOR hinders conversion of effector cells into T-memory cells when mTOR is expressed. Activated T cells are targeted by the 4-1BB aptamer, and after internalising 4-1BB, release Raptor siRNA into the cytoplasm of the cell (CD137). When memory T cells are being induced, mTOR is inhibited by the very existence of siRNA within the cell [92]. Dassie et al. proposed a specimen for an aptamer-siRNA chimaera that alters gene articulation. Dassie et al. identified an aptamer that binds to siRNA and causes tumours that express prostate-specific membrane antigen (PMSA) to recede [93].
Aptamers have the ability to conjugate with chemotherapy agents as well as therapeutic RNA or DNA. This interesting hypothesis might be extended to tumours that secrete MMP and/or kallikrein (klk). Use of an iRNA chimera aptamer as a delivery vehicle has also been described by Dassie et al. Moreover, Dassie et al. selected an RNA aptamer that targets genes that help prostate cancer cells remain viable and sends cytotoxic siRNA directly to the cancer cells [94]. The ability of aptamers to deliver active RNA to cells in vitro as well as in vivo was demonstrated for the first time in this work.
Many organizations have begun to use similar delivery strategies. An aptamer was used to administer DOX to HER2-positive cancerous breast cells according to Liu et al. DOX must bind to DNA to form the aptamer-DOX complex (Apt-DOX). Due to the structure of HB5, DOX is able to reach HER2-positive breast cancer cells and inhibit HER2-negative cells from drug uptake in vitro, resulting in less cytotoxicity [95].
Thiel et al. used RNA aptamers to deliver chemotherapy-sensitizing siRNAs to HER2-positive breast cancer cells. The goal was to find RNA aptamers that are specific for a certain type of cell that has cell-internalising properties. Thiel et al. developed a unique cell-based selection method [96]. These aptamers (anti-apoptotic genes) have siRNAs that are coupled covalently and target the Bcl-2 gene. Application of the HER2 aptamer-Bcl-2 siRNA pair specifically targeted HER2-positive cells, lowered the expression of the Bcl-2 gene, and made the cells more vulnerable to cisplatin therapy. These projects hold great potential because the disease is so unique and the treatment is difficult because of the unusual properties of malignant cells.
Targeted drug delivery (aptamer-based nanosystem)
An aptamer-based nanosystem contains various nanomaterials that have an important role in the application of biological medicine and analysis [97]. Nanomaterials have advantageous characteristics, such an ultra-micro size with large surface area and having high loading capacity over conventional theranostic strategies [Figure 4] [98].
Inorganic nanomedicine-based system
Silica-based nanoparticles: Silica nanoparticles are becoming an appropriate carrier in delivery of drugs in an inorganic system because silica nanoparticles provide the controlled drug release by changing pH, temperature, and redox and photochemical reactions [99]. When coupled with specific components, such as aptamers, silica nanoparticles improve the therapeutic effects of cancer treatment at a lower medication dose [100].
A new redox-responsive nanocontainer based on mesoporous silica nanoparticles (MSN) for targeted triplex cancer therapy was described by Cai et al. [101]. The AS1411 aptamer was customised for CytC-sealed MSNs [101]. Additionally, this mechanism may cause the S-S connections to dissolve, allowing DOX to be released specifically into the tumour cells. FRET-based two-photon MSNs were initially created by Tan et al. [102] in 2021 for targeted drug delivery and multiplexed intracellular imaging. By altering the doping ratio of the 3 dyes, the MSNs exhibit distinct two-photon multi-color fluorescence. Moreover, the anticancer medication, DOX, can be released from the DOX-loaded, aptamer-capped nanosystem and internalised into cancer cells with efficiency. An abundance of work has also been performed on gene-targeted delivery using aptamer-targeted MSNs, which can shield gene therapy molecules from nuclease degradation [103].
Carbon-based nanoparticles: Carbon nanomaterials functionalized with aptamers are perfect as nanoplatforms for cancer treatment and diagnostics [53]. Multi-functionality was recently produced by Wang et al. and demonstrated sustained release and heat-stimulating capabilities. This nanoparticle exhibits remarkable recognition ability for targeting MCF-7 breast cells due to the incorporation of MUC1 aptamers [104].
Gold-based nanoparticles: The biomedical field has widely accepted gold nanoparticles because of exceptional stability, low toxicity, high surface-to-volume ratio, and biological compatibility [105]. The use of aptamer-conjugated gold nanomaterials (Apt-AuNPs), which combine the unique benefits of aptamers with gold nanoparticles, has been extensively adopted in cancer treatment and diagnosis [106].
Yang et al. reported the use of apt-AuNPs in conjunction with graphene oxide for photothermal therapy of breast cancer [107]. Dou et al. [108] reported the use of aptamer-functionalized AuNPs-Fe3O4-GS capture probes for tracking circulating tumour cells in whole blood.
Organic-based system
Liposomal system: Liposomes are a promising drug delivery technology because of the high drug loading efficiency, low immunogencity, low toxicologic profile, and superior biocompatibility [109]. For targeted cancer gene therapy, a recent attempt to package the CRISPR/Cas9 complex into liposomes functionalized with an aptamer [110]. Liang et al. created a lipopolymer based on aptamers to transport CRISPR/Cas9 to specific tumours and control vascular endothelial growth factor (VEGFA) in osteosarcoma. The LC09 aptamer has the potential to enable the targeted delivery of CRISPR/Cas9 plasmids in this system, thereby reducing VEGFA expression and preventing lung metastasis and orthotopic osteosarcoma malignancy [111].
Micelle-based system: Micelle structure is another interesting class of organic nanomaterial based on aptamers. Because of the multivalent effect, this drug delivery method exhibits good aptamer binding to targets. A micelle-based system can therefore be developed for a wide range of bioapplications [112].
A cross-linked aptamer-lipid micelle system was created by Li et al. [113] in 2018 for target cell recognition that offers exceptional stability and specificity. In this simple method, an effective photoinduced polymerization process connected the aptamer and lipid segments to a methacrylamide branch [113].
Hydrogel-based system: Target-responsive DNA hydrogels are an aptamer-based organic nanomaterial system that has been applied extensively in pharmaceutical and biomedical fields. Target-responsive DNA hydrogels have outstanding mechanical properties and programmable features [114]. For possible drug release, the first adenosine-responsive hydrogel was created in 2008. This study used two oligonucleotide-incorporated polyacrylamides and carefully thought-out cross-linking oligonucleotides to create DNA nanohydrogels. The adenosine aptamer sequence was present in the DNA linker. The hydrogel will dissolve and the cross-links will break when the aptamer binds to target molecules that already contain adenosine. Accordingly, target-responsive drug release may be investigated using this method [115].
Challenges and overcoming methods in aptamer technology
Stability and nuclease resistance
Aptamers, especially aptamers based on RNA, are susceptible to degradation by nucleases. Enhancing stability and nuclease resistance is a significant challenge. Various modifications, such as chemical modifications or using modified nucleotides, can be used to increase the stability of aptamers. However, these alterations may impact aptamer binding properties and introduce additional complexities.
Identification of suitable aptamers from the aptamer library
As is well known, an aptamer sequence ultimately controls the 3-dimensional conformation, which in turn allows the aptamer to bind to its target. An aptamer library with highly varied sequences will include a wide variety of structurally unique aptamers. This diversity increases the likelihood of developing aptamers that bind tightly to desired targets and contrasts with proteins that utilize 20 different amino acids to present diverse functional groups, while nucleic acids rely on only 4 bases (adenine, guanine, cytosine, and thymine/uracil) and have fewer functional group options. Consequently, aptamers have functional groups added that mimic the side chains of amino acids to broaden the chemical range and confer protein-like properties [116].
A pioneer in expanding the variety of aptamer sequences in a library is SomaLogic, a top business in aptamer creation. New functional groups, such as naphthyl, benzyl, tryptamino, and isobutyl hydrophobic side chains, were added at the 5′ position of dUTP nucleotides. Slow off-rate changes in aptamers (SOMAmers), which were created utilizing these changed nucleotides. Therefore, aptamers modified with SOMAmer technology have been shown to improve aptamer selection compared to unmodified aptamers when targeting difficult proteins. The success rate rose from approximately 30% for standard aptamers to >90% for SOMAmers [117].
Target selection criteria for aptamers
Aptamers can be developed through an iterative process of selection and amplification from a large pool of randomized single-stranded DNA/RNA oligonucleotides (with >1015 random sequences) called SELEX. This method allows for internalisation of therapeutic compounds into cells when aptamers are chemically conjugated [118] to compounds as well as aptamer binding to extracellular targets on the cell surface [119], from which therapeutic aptamers have been produced. Although the selection process for creating aptamers for either use is fundamentally the same, a higher level of success must be achieved with the cell-internalising aptamers. A crucial factor that needs to be considered for cargo efficient biological activity is endosomal escape and delivery to the cytosol.
Filtration process through the kidney
For effective therapeutic use, it is necessary to overcome the phenomenon of renal filtration of small molecule medications. Aptamers experience this difficulty because of the diminutive size. The size of an aptamer with a mass of 6–30 kDa is approximately 5 nm [120]. Even with stabilizing backbone changes, unmodified aptamers supplied intravenously are prone to fast renal filtration excretion, which reduces circulation time.
Aptamers can be chemically modified by the addition of large compounds, such as polyethylene glycol (PEG), liposomes, proteins, and organic or inorganic nanomaterials. Aptamers can also be multimerized. These modifications enable the aptamers to achieve an overall mass that exceeds the molecular weight cut-off for glomerular filtration (30–50 kDa).
To circumvent this problem we used further modifications as follows.
PEGylation
One of the most popular techniques for preventing renal filtration of aptamers is PEGylation. PEG is highly lipophobic (i.e., hydrophilic), a flexible, uncharged polymer that is frequently used with medicinal medicines to increase medication efficacy, prolong circulation time, and decrease reticuloendothelial clearance [121].
The only aptamer-based drug that the FDA has currently approved is Macugen. Macugen utilizes PEGylation, a chemical strategy that grafts PEG modifications. PEG groups reduce clumping behaviour and heightens the solubility of conjugated parameters. For Macugen, PEGylation enables substantial enhancement of the plasma half-life (9.5–12.5 h) after intravenous delivery. Similarly, the vitreous humour half-life reached approximately 94 h upon subcutaneous administration owing to the PEG groups [122].
Chol-aptamer
An RNA aptamer with modifications to all pyrimidine nucleotides was previously shown to inhibit replication of hepatitis C virus (HCV). Researchers subsequently conjugated this aptamer to cholesterol, yielding a cholesterol-aptamer construct. When applied to cells, this cholesterol-linked aptamer exhibited efficient cellular uptake and halted HCV RNA amplification. Furthermore, systemic administration of the cholesterol-aptamer in a mouse model demonstrated good tolerability along with extended plasma circulation time [123].
Diacylglycerol (DAG) conjugation
DAG was conjugated at the 5′ end of the VEGF aptamer by Willis et al. [124]. After being added to a liposome bilayer, aptamers having enhanced anti-VEGF activity were generated by this DAG-aptamer combination, both in vitro and in vivo. This DAG-aptamer-liposome combination showed a much longer plasma residency time than an unaltered aptamer [124].
Toxicity
Aptamers have a significant advantage over antibodies with respect to limited or non-immunogenicity. Following efficacy and stability assessments, a critical next stage in medication testing is the evaluation of immunogenic potential, toxicity, and adverse effects. The aptamer-based drug, pegaptanib sodium (Macugen), which is used to treat wet age-related macular degeneration, did not show any signs of being harmful in preclinical tests [120].
Although there is insufficient clinical evidence of the cytotoxicity of aptamers, research using different animal models exhibited minimal or no evidence of cardiac toxicity, liver damage, or renal toxicity. Non-specific absorption causes cardiotoxicity, which is a side effect of the widely used anticancer drug, DOX. PEGylated or liposomal DOX has not been able to resolve this issue. In contrast to non-targeted administration, the aptamer-DOX conjugate is reported to minimize this cardiotoxicity and boost the efficacy of tumour suppression in vivo [125].
Targeting action of aptamer
Targeting the tumour-initiating cells that are developing and stem-like system
Most epithelia have an EpCAM, which is a glycosylated cell surface receptor that helps cells divide, migrate, and differentiate [126].
Most carcinomas, including the lung, breast, colon, pancreatic, and prostate, have high levels of EpCAM expression. This protein is also authorised as a biomarker for detecting circulating tumour cells and is implicated in the formation of stem-like tumour-initiating cells [38, 54, 127].
Focusing on intracranial brain cancer
The tight cerebrovascular endothelium that makes up the BBB, a physical barrier that poses a significant obstacle to the advancement of gene therapy drugs for brain tumours, the barrier hinders the flow of big macromolecules from the blood-to-the brain, including peptides and nucleic acids, which reduces the efficacy of treatments [128].
Jiang et al. recently explored transporting PTX, a highly potent chemotherapeutic for various cancers, including gliomas, across the BBB. Jiang et al. utilized AS1411, an aptamer targeting nucleolin receptors, as the guiding molecule [129]. Scientists made AS1411-PGG-PTX by connecting AS1411 and poly-(α-glutamyl-glutamine)-paclitaxel (PGG-PTX). This compound has two different functions. Incorporating PGG improved PTX solubility and diminished toxicity [130].
Addressing these challenges requires interdisciplinary collaboration and ongoing research efforts. Continued advances in aptamer technology have the potential to revolutionise various fields, ranging from theranostics to environmental monitoring and biosensing. By overcoming these challenges, aptamers can offer innovative solutions and contribute to the development of more efficient and targeted approaches in diverse applications.
Conclusion
Aptamer technology has developed tremendous potential as a tool centred on theranostics. Aptamer technology utilizes perspectives and various challenges in the design of aptamer technology for targeted cancer therapeutics. Aptamer technology is involved in the design of the aptamers and various fluorogenic aptamers, such as green aptamers, concluded the diagnostic approach. Various modifications, such as PEGylation and cholesterol-containing parameters, and many more to overcome challenges are particularly derived from the aptamer technology.
Various SELEX technologies targeted at selected molecules were presented in this article. SELEX technologies also involves various drugs that are used in chemotherapy studies, and we studied aptamers as biotherapy and chemotherapy agents and involved various types of aptamers.
We investigated the aptamer-exosome technology and how aptamer-exosome technology captures aptamers. We also investigated how exosomes diagnose aptamers and how exosomes can help with theranostic effects in specific systems.
Funding
No funding was received for publication of this review.
Conflicts of interest
The authors certify that there are no conflicts of interest with any financial organization regarding the material discussed in the manuscript.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Authors’ contributions
J.T. and M.Y. have made substantial contributions to the conception and design of the manuscript, acquisition, analysis, and interpretation of the data. A.S.T., and R.K.M. have participated in drafting the manuscript and revised it critically. All authors read and approved the final version of the manuscript. All authors contributed equally to the manuscript and read and approved the final version of the manuscript.
Graphical abstract
Oncology related design, strategy, and implementation of aptamer based nano-system in theranostics.
References
- Pereira RL, Nascimento IC, Santos AP, Ogusuku IEY, Lameu C, et al. Aptamers: novelty tools for cancer biology. Oncotarget 2018;9(42):26934-53. [PMID: 29928493 DOI: 10.18632/oncotarget.25260]
- Jin C, Wang K, Oppong-Gyebi A, Hu J. Application of nanotechnology in cancer diagnosis and therapy – a mini-review. Int J Med Sci 2020;17(18):2964-73. [PMID: 33173417 DOI: 10.7150/ijms.49801]
- Wei Z, Zhou Y, Wang R, Wang J, Chen Z. Aptamers as smart ligands for targeted drug delivery in cancer therapy. Pharmaceutics 2022;14(12):2561. [PMID: 36559056 DOI: 10.3390/pharmaceutics14122561]
- Oliveira M, Falato C, Cejalvo JM, Vila MM, Tolosa P, et al. Patritumab deruxtecan in untreated hormone receptor-positive/HER2-negative early breast cancer: final results from part A of the window-of-opportunity SOLTI TOT-HER3 pre-operative study. Ann Oncol 2023;34(8):670-80. [PMID: 37211044 DOI: 10.1016/j.annonc.2023.05.004]
- Agnello L, d’Argenio A, Nilo R, Fedele M, Camorani S, et al. Aptamer-based strategies to boost immunotherapy in TNBC. Cancers (Basel) 2023;15(7):2010. [PMID: 37046670 DOI: 10.3390/cancers15072010]
- Adachi T, Nakamura Y. Aptamers : a review of their chemical properties. Molecules 2019;24:4229. [PMID: 31766318 DOI: 10.3390/molecules24234229]
- Hori S, Herrera A, Rossi J, Zhou J. Current advances in aptamers for cancer diagnosis and therapy. Cancers (Basel) 2018;10(1):9. [PMID: 29301363 DOI: 10.3390/cancers10010009]
- Tran PH-L, Xiang D, Nguyen TNG, Tran TTD, Chen Q, et al. Aptamer-guided extracellular vesicle theranostics in oncology. Theranostics 2020;10(9):3849-66. [PMID: 32226524 DOI: 10.7150/thno.39706]
- Mayer G. The chemical biology of aptamers. Angew Chem Int Ed Engl 2009;48(15):2672-89. [PMID: 19319884 DOI: 10.1002/anie.200804643]
- Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov 2010;9(7):537-50. [PMID: 20592747 DOI: 10.1038/nrd3141]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990;249(4968):505-10. [PMID: 2200121 DOI: 10.1126/science.2200121]
- Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science 2000;287(5454):820-5. [PMID: 10657289 DOI: 10.1126/science.287.5454.820]
- Rajendran M, Ellington AD. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal Bioanal Chem 2008;390(4):1067-75. [PMID: 18049815 DOI: 10.1007/s00216-007-1735-8]
- Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A 2006;103(32):11838-43. [PMID: 16873550 DOI: 10.1073/pnas.0602615103]
- Tang Z, Shangguan D, Wang K, Shi H, Sefah K, et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem 2007;79(13):4900-7. [PMID: 17530817 DOI: 10.1021/ac070189y]
- Ng EWM, Shima DT, Calias P, Cunningham ET, Guyer DR, et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 2006;5(2):123-32. [PMID: 16518379 DOI: 10.1038/nrd1955]
- Shieh Y-A, Yang S-J, Wei M-F, Shieh M-J. Aptamer-based tumour-targeted drug delivery for photodynamic therapy. ACS Nano 2010;4(3):1433-42. [PMID: 20166743 DOI: 10.1021/nn901374b]
- Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin-targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res 2008;68(7):2358-65. [PMID: 18381443 DOI: 10.1158/0008-5472.CAN-07-5723]
- Xu J, Li H, Arumugam SS, Rong Y, Wang P, et al. A turn-on fluorescence sensor for rapid sensing of ATP based on luminescence resonance energy transfer between upconversion nanoparticles and Cy3 in vivo or vitro. Spectrochim Acta A Mol Biomol Spectrosc 2022;265:120341. [PMID: 34492515 DOI: 10.1016/j.saa.2021.120341]
- Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol 2009;86(3):151-64. [PMID: 19454272 DOI: 10.1016/j.yexmp.2009.01.004]
- Kumar Sharma T, Ramanathan R, Weerathunge P, Mohammadtaheri M, Kumar Daima H, et al. Aptamer-mediated “turn-off/turn-on” nanozyme activity of gold nanoparticles for kanamycin detection. Chem Commun 2014;50(100):15856-9. [PMID: 25331713 DOI: 10.1039/c4cc07275h]
- Das R, Dhiman A, Kapil A, Bansal V, Sharma TK. Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme. Anal Bioanal Chem 2019;411(6):1229-38. [PMID: 30637436 DOI: 10.1007/s00216-018-1555-z]
- Keppler D, Lin AW. Cervical cancer. In: Keppler D, Lin AW, editors. Cervical cancer: methods and protocols (Methods in molecular biology; vol. 1249). New York, NY: Springer New York; 2015. pp. 1-406. Available from: https://link.springer.com/10.1007/978-1-4939-2013-6.
- Bala J, Chinnapaiyan S, Dutta RK, Unwalla H. Aptamers in HIV research diagnosis and therapy. RNA Biol 2018;15(3):327-37. [PMID: 29431588 DOI: 10.1080/15476286.2017.1414131]
- Yadav AK, Verma D, Chaudhary N, Kumar A, Solanki PR. Aptamer based switches: a futuristic approach for Helicobacter pylori detection. Mater Lett 2022;308:131239. [DOI: 10.1016/j.matlet.2021.131239]
- Palareti G, Legnani C, Cosmi B, Antonucci E, Erba N, et al. Comparison between different D-Dimer cutoff values to assess the individual risk of recurrent venous thromboembolism: analysis of results obtained in the DULCIS study. Int J Lab Hematol 2016;38(1):42-9. [PMID: 26362346 DOI: 10.1111/ijlh.12426]
- Robertson DL, Joyce GF. Selection in-vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990;344:467-8. [PMID: 1690861 DOI: 10.1038/344467a]
- Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science 2011;333(6042):642-6. [PMID: 21798953 DOI: 10.1126/science.1207339]
- Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc 2014;136(46):16299-308. [PMID: 25337688 DOI: 10.1021/ja508478x]
- Dolgosheina EV, Jeng SCY, Panchapakesan SSS, Cojocaru R, Chen PSK, et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol 2014;9(10):2412-20. [PMID: 25101481 DOI: 10.1021/cb500499x]
- Song W, Filonov GS, Kim H, Hirsch M, Li X, et al. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat Chem Biol 2017;13(11):1187-94. [PMID: 28945233 DOI: 10.1038/nchembio.2477]
- Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, et al. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol 2014;10(8):686-91. [PMID: 24952597 DOI: 10.1038/nchembio.1561]
- TrachmanIii RJ, Demeshkina NA, Lau MWL, Panchapakesan SSS, Jeng SCY, et al. Structural basis for high-affinity fluorophore binding and activation by RNA Mango. Nat Chem Biol 2017;13(7):807-13. [PMID: 28553947 DOI: 10.1038/nchembio.2392]
- Yoon S, Rossi JJ. Targeted molecular imaging using aptamers in cancer. Pharmaceuticals 2018;11(3):1-16. [PMID: 30029472 DOI: 10.3390/ph11030071]
- Tkach M, Kowal J, Théry C. Why the need and how to approach the functional diversity of extracellular vesicles. Philos Trans R Soc B Biol Sci 2018;373(1737). [PMID: 29158309 DOI: 10.1098/rstb.2016.0479]
- Zijlstra A, Di Vizio D. Size matters in nanoscale communication. Nat Cell Biol 2018;20(3):228-30. [PMID: 29476154 DOI: 10.1038/s41556-018-0049-8]
- Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol 1967;13(3):269-88. [PMID: 6025241 DOI: 10.1111/j.1365-2141.1967.tb08741.x]
- Soares RP, Xander P, Costa AO, Marcilla A, Menezes-Neto A, et al. Highlights of the São Paulo ISEV workshop on extracellular vesicles in cross-kingdom communication. J Extracell Vesicles 2017;6(1). [PMID: 30044885 DOI: 10.1080/20013078.2017.1407213]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, et al. Reassessment of exosome composition. Cell 2019;177(2):428-45.e18. [PMID: 30951670 DOI: 10.1016/j.cell.2019.02.029]
- Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci 2019;9(1):1-18. [PMID: 30815248 DOI: 10.1186/s13578-019-0282-2]
- Im H, Shao H, Park YI, Peterson VM, Castro CM, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol 2014;32(5):490-5. [PMID: 24752081 DOI: 10.1038/nbt.2886]
- Shao H, Im H, Castro CM, Breakefield X, Weissleder R, et al. New technologies for analysis of extracellular vesicles. Chem Rev 2018;118(4):1917-50. [PMID: 29384376 DOI: 10.1021/acs.chemrev.7b00534]
- Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res 2017;66:30-41. [PMID: 28342835 DOI: 10.1016/j.plipres.2017.03.001]
- Jiang L, Gu Y, Du Y, Liu J. Exosomes: diagnostic biomarkers and therapeutic delivery vehicles for cancer. Mol Pharm 2019;16:3333-49. [PMID: 31241965 DOI: 10.1021/acs.molpharmaceut.9b00409]
- Chen G, Huang AC, Zhang W, Zhang G, Wu M, et al. S41586-018-0392-8. Nature 2018;560:382-6. [PMID: 30089911 DOI: 10.1038/s41586-018-0392-8]
- Hurwitz SN, Meckes DG. Extracellular vesicle integrins distinguish unique cancers. Proteomes 2019;7(2):14. [PMID: 30979041 DOI: 10.3390/proteomes7020014]
- Jan AT, Malik MA, Rahman S, Yeo HR, Lee EJ, et al. Perspective insights of exosomes in neurodegenerative diseases: a critical appraisal. Front Aging Neurosci 2017;9(SEP):1-8. [PMID: 29033828 DOI: 10.3389/fnagi.2017.00317]
- Tran PHL, Wang T, Yin W, Tran TTD, Barua HT, et al. Development of a nanoamorphous exosomal delivery system as an effective biological platform for improved encapsulation of hydrophobic drugs. Int J Pharm 2019;566(June):697-707. [PMID: 31207280 DOI: 10.1016/j.ijpharm.2019.06.028]
- Gobbo J, Marcion G, Cordonnier M, Dias AMM, Pernet N, et al. Restoring anticancer immune response by targeting tumour-derived exosomes with a HSP70 peptide aptamer. J Natl Cancer Inst 2016;108(3):1-11. [PMID: 26598503 DOI: 10.1093/jnci/djv330]
- Murakami K, Zhao J, Yamasaki K, Miyagishi M. Biochemical and structural features of extracellular vesicle-binding RNA aptamers. Biomed Rep 2017;6(6):615-26. [PMID: 28584632 DOI: 10.3892/br.2017.899]
- Zhao L, Huang Y, Dong Y, Han X, Wang S, et al. Aptamers and aptasensors for highly specific recognition and sensitive detection of marine biotoxins: recent advances and perspectives. Toxins (Basel) 2018;10(11):427. [PMID: 30366456 DOI: 10.3390/toxins10110427]
- Jin D, Yang F, Zhang Y, Liu L, Zhou Y, et al. ExoAPP: exosome-oriented, aptamer nanoprobe-enabled surface proteins profiling and detection. Anal Chem 2018;90(24):14402-11. [PMID: 30350954 DOI: 10.1021/acs.analchem.8b03959]
- Wan Y, Wang L, Zhu C, Zheng Q, Wang G, et al. Aptamer-conjugated extracellular nanovesicles for targeted drug delivery. Cancer Res 2018;78(3):798-808. [PMID: 29217761 DOI: 10.1158/0008-5472.CAN-17-2880]
- Pi F, Binzel DW, Lee TJ, Li Z, Sun M, et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol 2018;13(1):82-9. [PMID: 29230043 DOI: 10.1038/s41565-017-0012-z]
- Mairal T, Cengiz Özalp V, Lozano Sánchez P, Mir M, Katakis I, et al. Aptamers: molecular tools for analytical applications. Anal Bioanal Chem 2008;390(4):989-1007. [PMID: 17581746 DOI: 10.1007/s00216-007-1346-4]
- Bhalla N, Jolly P, Formisano N, Estrela P. Introduction to biosensors. Essays Biochem 2016;60(1):1-8. [PMID: 27365030 DOI: 10.1042/EBC20150001]
- Numnuam A, Chumbimuni-Torres KY, Xiang Y, Bash R, Thavarungkul P, et al. Aptamer-based potentiometric measurements of proteins using ion-selective microelectrodes. Anal Chem 2008;80(3):707-12. [PMID: 18184015 DOI: 10.1021/ac701910r]
- Ikebukuro K, Kiyohara C, Sode K. Electrochemical detection of protein using a double aptamer sandwich. Anal Lett 2004;37(14):2901-9. [DOI: 10.1081/AL-200035778]
- Pavlov V, Xiao Y, Shlyahovsky B, Willner I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J Am Chem Soc 2004;126(38):11768-9. [PMID: 15382892 DOI: 10.1021/ja046970u]
- Zhang JJ, Cheng FF, Zheng TT, Zhu JJ. Versatile aptasensor for electrochemical quantification of cell surface glycan and naked-eye tracking glycolytic inhibition in living cells. Biosens Bioelectron 2017;89:937-45. [PMID: 27818049 DOI: 10.1016/j.bios.2016.09.087]
- Xu Y, Cheng G, He P, Fang Y. A review: electrochemical aptasensors with various detection strategies. Electroanalysis 2009;21(11):1251-9. [DOI: 10.1002/elan.200804561]
- Wang K, He MQ, Zhai FH, He RH, Yu YL. A novel electrochemical biosensor based on polyadenine modified aptamer for label-free and ultrasensitive detection of human breast cancer cells. Talanta 2017;166:87-92. [PMID: 28213264 DOI: 10.1016/j.talanta.2017.01.052]
- Davis KA, Abrams B, Lin Y, Jayasena SD. Use of a high affinity DNA ligand in flow cytometry. Nucleic Acids Res 1996;24(4):702-6. [PMID: 8604313 DOI: 10.1093/nar/24.4.702]
- Herr JK, Smith JE, Medley CD, Shangguan D, Tan W. Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal Chem 2006;78(9):2918-24. [PMID: 16642976 DOI: 10.1021/ac052015r]
- Chen X, Estévez MC, Zhu Z, Huang YF, Chen Y, et al. Using aptamer-conjugated fluorescence resonance energy transfer nanoparticles for multiplexed cancer cell monitoring. Anal Chem 2009;81(16):7009-14. [PMID: 19572554 DOI: 10.1021/ac9011073]
- Freeman R, Girsh J, Jou AF, Ho JAA, Hug T, et al. Optical aptasensors for the analysis of the vascular endothelial growth factor (VEGF). Anal Chem 2012;84(14):6192-8. [PMID: 22746189 DOI: 10.1021/ac3011473]
- Cho H, Yeh EC, Sinha R, Laurence TA, Bearinger JP, et al. Single-step nanoplasmonic VEGF 165 aptasensor for early cancer diagnosis. ACS Nano 2012;6(9):7607-14. [PMID: 22880609 DOI: 10.1021/nn203833d]
- Xu H, Wu D, Li CQ, Lu Z, Liao XY, et al. Label-free colorimetric detection of cancer related gene based on two-step amplification of molecular machine. Biosens Bioelectron 2017;90(November 2016):314-20. [PMID: 27936442 DOI: 10.1016/j.bios.2016.12.003]
- Ahirwar R, Nahar P. Development of a label-free gold nanoparticle-based colorimetric aptasensor for detection of human estrogen receptor alpha. Anal Bioanal Chem 2016;408(1):327-32. [PMID: 26476919 DOI: 10.1007/s00216-015-9090-7]
- Shahriari M, Kesharwani P, Sahebkar A. Aptamer-based theranostic approaches for treatment of cancer. In: Kesharwani P, editor. Aptamers engineered nanocarriers for cancer therapy (Woodhead publishing series in biomaterials). Elsevier; 2023. pp. 433-54. Available from: https://www.sciencedirect.com/science/article/pii/B9780323858816000166.
- Li X, Zhou H, Yang L, Du G, Pai-Panandiker AS, et al. Enhancement of cell recognition in-vitro by dual-ligand cancer targeting gold nanoparticles. Biomaterials 2011;32(10):2540-5. [PMID: 21232787 DOI: 10.1016/j.biomaterials.2010.12.031]
- Feng C, Rui M, Shen H, Xin Y, Zhang J, et al. Tumour-specific delivery of doxorubicin through conjugation of pH-responsive peptide for overcoming drug resistance in cancer. Int J Pharm 2017;528(1-2):322-33. [PMID: 28606508 DOI: 10.1016/j.ijpharm.2017.06.022]
- Bates PJ, Reyes-Reyes EM, Malik MT, Murphy EM, O’Toole MG, et al. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: uses and mechanisms. Biochim Biophys Acta Gen Subj 2017;1861(5):1414-28. [PMID: 28007579 DOI: 10.1016/j.bbagen.2016.12.015]
- Liao ZX, Chuang EY, Lin CC, Ho YC, Lin KJ, et al. An AS1411 aptamer-conjugated liposomal system containing a bubble-generating agent for tumour-specific chemotherapy that overcomes multi-drug resistance. J Control Release 2015;208:42-51. [PMID: 25637705 DOI: 10.1016/j.jconrel.2015.01.032]
- Trinh TL, Zhu G, Xiao X, Puszyk W, Sefah K, et al. A synthetic aptamer-drug adduct for targeted liver cancer therapy. PLoS One 2015;10(11):1-17. [PMID: 26523833 DOI: 10.1371/journal.pone.0136673]
- Atabi F, Gargari SLM, Hashemi M, Yaghmaei P. Doxorubicin loaded DNA aptamer linked myristilated chitosan nanogel for targeted drug delivery to prostate cancer. Iran J Pharm Res 2017;16(1):35-49. [PMID: 28496460]
- Sampath P, Rhines LD, DiMeco F, Tyler BM, Park MC, et al. Interstitial docetaxel (Taxotere), ccarmustine and ccombined interstitial ttherapy: a novel treatment for experimental malignant glioma. J Neurooncol 2006;80(1):9-17. [PMID: 16636748 DOI: 10.1007/s11060-006-9159-4]
- Gao H, Qian J, Cao S, Yang Z, Pang Z, et al. Precise glioma targeting of and penetration by aptamer and peptide dual-functioned nanoparticles. Biomaterials 2012;33(20):5115-23. [PMID: 22484043 DOI: 10.1016/j.biomaterials.2012.03.058]
- Zhao M, Liang C, Li A, Chang J, Wang H, et al. Magnetic paclitaxel nanoparticles inhibit glioma growth and improve the survival of rats bearing glioma xenografts. Anticancer Res 2010;30(6):2217-23. [PMID: 20651372]
- Taghavi S, Ramezani M, Alibolandi M, Abnous K, Taghdisi SM. Chitosan-modified PLGA nanoparticles tagged with 5TR1 aptamer for in vivo tumour-targeted drug delivery. Cancer Lett 2017;400:1-8. [PMID: 28412238 DOI: 10.1016/j.canlet.2017.04.008]
- Zhao X, Lis JT, Shi H. A systematic study of the features critical for designing a high avidity multivalent aptamer. Nucleic Acid Ther 2013;23(3):238-42. [PMID: 23550551 DOI: 10.1089/nat.2012.0410]
- Lozano T, Soldevilla MM, Casares N, Villanueva H, Bendandi M, et al. Targeting inhibition of Foxp3 by a CD28 2’-Fluro oligonucleotide aptamer conjugated to P60-peptide enhances active cancer immunotherapy. Biomaterials 2016;91:73-80. [PMID: 26999456 DOI: 10.1016/j.biomaterials.2016.03.007]
- Cortes-Dericks L, Schmid RA. CD44 and its ligand hyaluronan as potential biomarkers in malignant pleural mesothelioma: evidence and perspectives. Respir Res 2017;18(1):1-12. [PMID: 28403901 DOI: 10.1186/s12931-017-0546-5]
- Phua LC, Chue XP, Koh PK, Cheah PY, Ho HK, et al. Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol Ther 2014;15(4):389-97. [PMID: 24424155 DOI: 10.4161/cbt.27625]
- Holst S, Stavenhagen K, Balog CIA, Koeleman CAM, McDonnell LM, et al. Investigations on aberrant glycosylation of glycosphingolipids in colorectal cancer tissues using liquid chromatography and matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS). Mol Cell Proteomics 2013;12(11):3081-93. [PMID: 23878401 DOI: 10.1074/mcp.M113.030387]
- Manna SK, Tanaka N, Krausz KW, Haznadar M, Xue X, et al. Biomarkers of coordinate metabolic reprogramming in colorectal tumours in mice and humans. Gastroenterology 2014;146(5):1313-24. [PMID: 24440673 DOI: 10.1053/j.gastro.2014.01.017]
- Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, et al. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol 2007;2:751-60. [PMID: 18654426 DOI: 10.1038/nnano.2007.387]
- Lammers T, Kiessling F, Hennink WE, Storm G. Drug targeting to tumours: principles, pitfalls and (pre-) clinical progress. J Control Release 2012;161(2):175-87. [PMID: 21945285 DOI: 10.1016/j.jconrel.2011.09.063]
- Xuan W, Peng Y, Deng Z, Peng T, Kuai H, et al. A basic insight into aptamer-drug conjugates (ApDCs). Biomaterials 2018;182:216-26. [PMID: 30138784 DOI: 10.1016/j.biomaterials.2018.08.021]
- Chen K, Liu B, Yu B, Zhong W, Lu Y, et al. Advances in the development of aptamer-drug conjugates for targeted drug delivery. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2017;9(3). [PMID: 27800663 DOI: 10.1002/wnan.1438]
- Huang YF, Shangguan D, Liu H, Phillips JA, Zhang X, et al. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumour cells. ChemBioChem 2009;10(5):862-8. [PMID: 19253922 DOI: 10.1002/cbic.200800805]
- Berezhnoy A, Castro I, Levay A, Malek TR, Gilboa E. Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity. J Clin Invest 2014;124(1):188-97. [PMID: 24292708 DOI: 10.1172/JCI69856]
- Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumours. Nat Biotechnol 2009;27(9):839-46. [PMID: 19701187 DOI: 10.1038/nbt.1560]
- Lupold SE, Hicke BJ, Lin Y. Correction: identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res 2002;62:4029-33. Erratum in: Cancer Res 2012;72(15):3887. [PMID: 12124337]
- Liu Z, Duan JH, Song YM, Ma J, Wang FD, et al. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in-vitro. J Transl Med 2012;10(1):1-10. [PMID: 22817844 DOI: 10.1186/1479-5876-10-148]
- Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, et al. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res 2012;40(13):6319-37. [PMID: 22467215 DOI: 10.1093/nar/gks294]
- Li SL, Jiang P, Jiang FL, Liu Y. Recent advances in nanomaterial-based nanoplatforms for chemodynamic cancer therapy. Adv Funct Mater 2021;31(22):1-26. [DOI: 10.1002/adfm.202100243]
- Dong Y, Yao C, Zhu Y, Yang L, Luo D, et al. Dna functional materials assembled from branched dna: design, synthesis, and applications. Chem Rev 2020;120(17):9420-81. [PMID: 32672036 DOI: 10.1021/acs.chemrev.0c00294]
- Fu Z, Xiang J. Aptamer-functionalized nanoparticles in targeted delivery and cancer therapy. Int J Mol Sci 2020;21(23):1-39. [PMID: 33266216 DOI: 10.3390/ijms21239123]
- Vandghanooni S, Barar J, Eskandani M, Omidi Y. Aptamer-conjugated mesoporous silica nanoparticles for simultaneous imaging and therapy of cancer. TrAC Trends Anal Chem 2020;123:115759. [DOI: 10.1016/j.trac.2019.115759]
- Zhang B, Luo Z, Liu J, Ding X, Li J, et al. Cytochrome c end-capped mesoporous silica nanoparticles as redox-responsive drug delivery vehicles for liver tumour-targeted triplex therapy in-vitro and in vivo. J Control Release 2014;192:192-201. [PMID: 25034575 DOI: 10.1016/j.jconrel.2014.06.037]
- Wu YX, Zhang D, Hu X, Peng R, Li J, Zhang X, et al. Multicolor two-photon nanosystem for multiplexed intracellular imaging and targeted cancer therapy. Angew Chem Int Ed Engl 2021;60(22):12569-76. [PMID: 33739576 DOI: 10.1002/anie.202103027]
- Zhang P, Cheng F, Zhou R, Cao J, Li J, et al. DNA-hybrid-gated multi-functional mesoporous silica nanocarriers for dual-targeted and microRNA-responsive controlled drug delivery. Angew Chem Int Ed Engl 2014;53(9):2371-5. [PMID: 24470397 DOI: 10.1002/anie.201308920]
- Wang X, Han Q, Yu N, Li J, Yang L, et al. Aptamer-conjugated graphene oxide-gold nanocomposites for targeted chemo-photothermal therapy of cancer cells. J Mater Chem B 2015;3(19):4036-42. [PMID: 32262625 DOI: 10.1039/c5tb00134j]
- Yang L, Tseng YT, Suo G, Chen L, Yu J, et al. Photothermal therapeutic response of cancer cells to aptamer-gold nanoparticle-hybridized graphene oxide under NIR illumination. ACS Appl Mater Interfaces 2015;7(9):5097-106. [PMID: 25705789 DOI: 10.1021/am508117e]
- Nooranian S, Mohammadinejad A, Mohajeri T, Aleyaghoob G, Kazemi Oskuee R. Biosensors based on aptamer-conjugated gold nanoparticles: a review. Biotechnol Appl Biochem 2022;69(4):1517-34. [PMID: 34269486 DOI: 10.1002/bab.2224]
- Yang X, Yang M, Pang B, Vara M, Xia Y. Gold nanomaterials at work in biomedicine. Chem Rev 2015;115(19):10410-88. [PMID: 26293344 DOI: 10.1021/acs.chemrev.5b00193]
- Dou B, Xu L, Jiang B, Yuan R, Xiang Y. Aptamer-functionalized and gold nanoparticle array-decorated magnetic graphene nanosheets enable multiplexed and sensitive electrochemical detection of rare circulating tumour cells in whole blood. Anal Chem 2019;91(16):10792-9. [PMID: 31310099 DOI: 10.1021/acs.analchem.9b02403]
- Moosavian SA, Sahebkar A. Aptamer-functionalized liposomes for targeted cancer therapy. Cancer Lett 2019;448;144-54. [PMID: 30763718 DOI: 10.1016/j.canlet.2019.01.045]
- Gong Y, Tian S, Xuan Y, Zhang S. Lipid and polymer mediated CRISPR/Cas9 gene editing. J Mater Chem B 2020;8(20):4369-86. [PMID: 32364557 DOI: 10.1039/d0tb00207k]
- Liang C, Li F, Wang L, Zhang ZK, Wang C, et al. Tumour cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials 2017;147:68-85. [PMID: 28938163 DOI: 10.1016/j.biomaterials.2017.09.015]
- Wu Y, Sefah K, Liu H, Wang R, Tan W. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci U S A 2010;107(1):5-10. [PMID: 20080797 DOI: 10.1073/pnas.0909611107]
- Li X, Figg CA, Wang R, Jiang Y, Lyu Y, et al. Cross-linked aptamer–lipid micelles for excellent stability and specificity in target cell recognition. Angew Chem Int Ed Engl 2018;57(36):11589-93. [PMID: 30079455 DOI: 10.1002/anie.201804682]
- Li J, Mo L, Lu CH, Fu T, Yang HH, et al. Functional nucleic acid-based hydrogels for bioanalytical and biomedical applications. Chem Soc Rev 2016;45(5):1410-31. [PMID: 26758955 DOI: 10.1039/c5cs00586h]
- Yang H, Liu H, Kang H, Tan W. Engineering target-responsive hydrogels based on aptamer-target interactions. J Am Chem Soc 2008;130(20):6320-1. [PMID: 18444626 DOI: 10.1021/ja801339w]
- Eaton BE. The joys of in-vitro selection : chemically dressing oligonucleotides to satiate protein targets. Curr Opin Chem Biol 1997;10-6. [PMID: 9667830 DOI: 10.1016/s1367-5931(97)80103-2]
- Gold L, Ayers D, Bertino J, Bock C, Bock A, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 2010;5(12):e15004. [PMID: 21165148 DOI: 10.1371/journal.pone.0015004]
- Yoon S, Rossi JJ. Aptamers: uptake mechanisms and intracellular applications. Adv Drug Deliv Rev 2018;134:22-35. [PMID: 29981799 DOI: 10.1016/j.addr.2018.07.003]
- Vinores SA. Pegaptanib in the treatment of wet, age-related macular degeneration. Int J Nanomedicine 2006;1(3):263-8. [PMID: 17717967]
- Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov 2017;16(3):181-202. [PMID: 27807347 DOI: 10.1038/nrd.2016.199]
- Yang Q, Lai SK. Anti-PEG immunity: emergence, characteristics, and unaddressed questions. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2015;7(5):655-77. [PMID: 25707913 DOI: 10.1002/wnan.1339]
- Tucker CE, Chen LS, Judkins MB, Farmer JA, Gill SC, et al. Detection and plasma pharmacokinetics of an anti-vascular endothelial growth factor oligonucleotide-aptamer (NX1838) in rhesus monkeys. J Chromatogr B Biomed Sci Appl 1999;732(1):203-12. [PMID: 10517237 DOI: 10.1016/s0378-4347(99)00285-6]
- Lee CH, Lee SH, Kim JH, Noh YH, Noh GJ, et al. Pharmacokinetics of a cholesterol-conjugated aptamer against the hepatitis C virus (HCV) NS5B protein. Mol Ther Nucleic Acids 2015;4(10):e254. [PMID: 26440598 DOI: 10.1038/mtna.2015.30]
- Willis MC, Collins B, Zhang T, Green LS, Sebesta DP, et al. Liposome-anchored vascular endothelial growth factor aptamers. Bioconjug Chem 1998;9(5):573-82. [PMID: 9736491 DOI: 10.1021/bc980002x]
- Dou XQ, Wang H, Zhang J, Wang F, Xu GL, et al. Aptamer-drug conjugate: targeted delivery of doxorubicin in a HER3 aptamer-functionalized liposomal delivery system reduces cardiotoxicity. Int J Nanomedicine 2018;13:763-76. [PMID: 29440899 DOI: 10.2147/IJN.S149887]
- Maetzel D, Denzel S, Mack B, Canis M, Went P, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol 2009;11(2):162-71. [PMID: 19136966 DOI: 10.1038/ncb1824]
- Imrich S, Hachmeister M, Gires O. EpCAM and its potential role in tumour-initiating cells. Cell Adh Migr 2012;6(1):30-8. [PMID: 22647938 DOI: 10.4161/cam.18953]
- Barar J, Rafi MA, Pourseif MM, Omidi Y. Blood-brain barrier transport machineries and targeted therapy of brain diseases. BioImpacts 2016;6(4):225-48. [PMID: 28265539 DOI: 10.15171/bi.2016.30]
- Luo Z, Yan Z, Jin K, Pang Q, Jiang T, et al. Precise glioblastoma targeting by AS1411 aptamer-functionalized poly (L-γ-glutamylglutamine)–Paclitaxel nanoconjugates. J Colloid Interface Sci 2017;490:783-96. [PMID: 27988470 DOI: 10.1016/j.jcis.2016.12.004]
- Yang D, Van S, Jiang X, Yu L. Novel free Paclitaxel-loaded poly(L-γ-glutamylglutamine)-paclitaxel nanoparticles. Int J Nanomedicine 2011;6(1):85-91. [PMID: 21289985 DOI: 10.2147/IJN.S15839]
- Stoltenburg R, Reinemann C, Strehlitz B. SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 2007;24(4):381-403. [PMID: 17627883 DOI: 10.1016/j.bioeng.2007.06.001]
- Ma L, Liu J. Catalytic nucleic acids: biochemistry, chemical biology, biosensors, and nanotechnology. iScience 2020;23(1):100815. [PMID: 31954323 DOI: 10.1016/j.isci.2019.100815]
- Li J, Green AA, Yan H, Fan C. Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat Chem 2017;9(11):1056-67. [PMID: 29064489 DOI: 10.1038/nchem.2852]
- Zhu G, Qiu L, Meng H, Mei L, Tan W. Aptamers-guided DNA nanomedicine for cancer theranostics. In: Tan W, Fang X, editors. Aptamers selected by cell-SELEX for theranostics. Berlin, Heidelberg: Springer; 2015. pp. 111-37. [DOI: 10.1007/978-3-662-46226-3_6]
- Kim M, Kim DM, Kim KS, Jung W, Kim DE. Applications of cancer cell-specific aptamers in targeted delivery of anticancer therapeutic agents. Molecules 2018;23(4):1-20. [PMID: 29617327 DOI: 10.3390/molecules23040830]
- Zhu C, Yang G, Ghulam M, Li L, Qu F. Evolution of multi-functional capillary electrophoresis for high-efficiency selection of aptamers. Biotechnol Adv 2019;37(8):107432. [PMID: 31437572 DOI: 10.1016/j.biotechadv.2019.107432]
- Wang T, Yin W, AlShamaileh H, Zhang Y, Tran PHL, et al. A detailed protein-SELEX protocol allowing visual assessments of individual steps for a high success rate. Hum Gene Ther Methods 2019;30(1):1-16. [PMID: 30700146 DOI: 10.1089/hgtb.2018.237]
- Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, et al. 2′-fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem 1998;273(32):20556-67. [PMID: 9685413 DOI: 10.1074/jbc.273.32.20556]
- Murphy C, Muscat A, Ashley D, Mukaro V, West L, et al. Tailored NEOadjuvant epirubicin, cyclophosphamide and nanoparticle albumin-bound paclitaxel for breast cancer: the phase II NEONAB trial–clinical outcomes and molecular determinants of response. PLoS One 2019;14(2):1-20. [PMID: 30763338 DOI: 10.1371/journal.pone.0210891]
- Murphy C, Mukaro V, Tobler R, Asher R, Gibbs E, et al. Evaluating the role of magnetic resonance imaging post-neoadjuvant therapy for breast cancer in the NEONAB trial. Intern Med J 2018;48(6):699-705. [PMID: 28869790 DOI: 10.1111/imj.13617]
- Libutti SK, Choyke PL, Alexander HR, Glenn G, Bartlett DL, et al. Clinical and genetic analysis of patients with pancreatic neuroendocrine tumours associated with von Hippel-Lindau disease. Surgery 2000;128(6):1022-8. [PMID: 11114638 DOI: 10.1067/msy.2000.110239]
- Rosenberg JE, Bambury RM, Van Allen EM, Drabkin HA, Lara PN, et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest New Drugs 2014;32(1):178-87. [PMID: 24242861 DOI: 10.1007/s10637-013-0045-6]
- Steurer M, Montillo M, Scarfò L, Mauro FR, Andel J, et al. Olaptesed pegol (NOX-A12) with bendamustine and rituximab: a phase IIa study in patients with relapsed/refractory chronic lymphocytic leukaemia. Haematologica 2019;104(10):2053-60. [PMID: 31097627 DOI: 10.3324/haematol.2018.205930]
- Park EJ, Choi J, Lee KC, Na DH. Emerging PEGylated non-biologic drugs. Expert Opin Emerg Drugs 2019;24(2):107-19. [PMID: 30957581 DOI: 10.1080/14728214.2019.1604684]
- National Library of Medicine (U.S.). The clinical application of 68Ga labeled ssDNA aptamer Sgc8 in healthy volunteers and colorectal patients. Identifier NCT03385148. 2017. Available from: https://clinicaltrials.gov/study/NCT03385148. [Accessed on 23rd Feb 2024].
- National Library of Medicine (U.S.). Study of AM003 in patients with locally advanced and metastatic solid tumours. Identifier NCT06258330. 2022. Available from: https://clinicaltrials.gov/study/NCT06258330. [Accessed on 23rd Feb 2024].