Voice Series: Interview with Professor Dr. Xin Gao, Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences
Executive Editor of BIOI
Published Online: November 22 2023
Cite this paper:
Phei Er Saw. Voice Series: Interview with Professor Dr. Xin Gao, Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences. BIO Integration 2023; 4(4): 141–144.
DOI: 10.15212/bioi-2023-0011. Available at: https://bio-integration.org/
Download citation
© 2023 The Authors. This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See https://bio-integration.org/copyright-and-permissions/
Foreword
In this Voice Series, we interviewed Professor Xin Gao. His research focuses on the breakthrough of key technologies, including front end multimode imaging technology, pre-operative planning and intra-operative navigation technology, and prognostic assessment techniques based on large amounts of clinical data and artificial intelligence. The purpose of Professor Gao’s research is to create integrated solutions for interventional diagnosis and therapy. Professor Gao has recently presided over 10 scientific research projects, published greater than 70 scientific papers, and obtained more than 18 invention patents.
EE: Thank you Professor Gao for being so gracious in accepting an interview invitation from BIOI. Can you tell us more about your background and your current research interests?
XG: Certainly, thank you for having me. My name is Xin Gao and I am a Professor at Suzhou Institute of Biomedical Engineering and Technology at the Chinese Academy of Sciences. I was affiliated was Zhejiang University, where I gained experience in interventional diagnosis and therapy. In my current research, I am interested in using advanced AI techniques to analyse large-scale omics data, such as radiomics, pathomics, and genomics, to identify biomarkers and molecular signatures associated with diseases. By integrating multiple omics data sets, I hope to develop more accurate disease diagnoses, prognoses, and treatment strategies, which ultimately lead to improved patient outcomes.
EE: Based on your background, we understand that you are currently doing groundbreaking research in AI-related fields. What captured your interest and motivated you to venture into this field in the beginning?
XG: My interest in AI-related fields began about 6 years ago when our group was working on tumour segmentation and predicting tumour treatment responses using medical images. We discovered that emerging AI technologies significantly outperformed traditional methods. I was fascinated by the potential of AI and machine learning algorithms to solve complex problems in healthcare. I was particularly drawn to the potential of AI to revolutionize medicine and healthcare by enabling faster and more accurate diagnoses, more effective treatments, and ultimately better patient outcomes. The vast amount of data being generated in healthcare, coupled with the increasing availability of powerful computational resources, make this an exciting time to be working in this field.
As I delved deeper into this area, I became more interested in the application of AI in precision medicine, specifically in the analysis of large-scale omics data sets. The ability to integrate multiple omics data types and use machine learning algorithms to extract meaningful insight from these data sets has tremendous potential to transform our understanding of disease and improve patient care.
Overall, I am driven by the potential of AI to have a real impact on people’s lives, and I am excited to be at the forefront of this rapidly evolving field.
EE: Could you share with us some of your major breakthroughs during your years of research in this field?
XG: Sure, I would be happy to. In my years of research in AI-related fields, I have had the opportunity to work on several projects that have led to significant breakthroughs. Here are a few representative papers that highlight some of these breakthroughs:
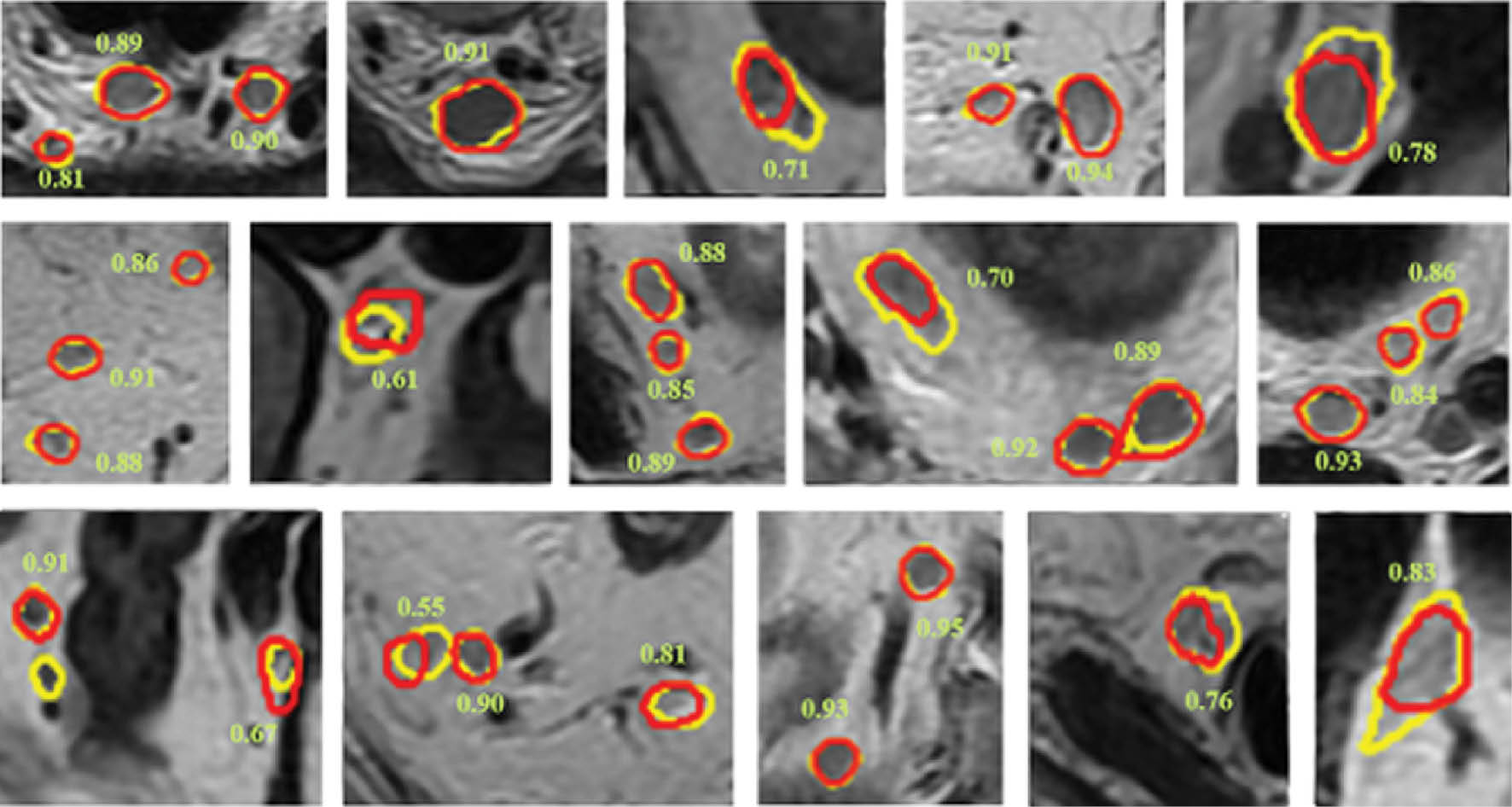
- “Deep learning–based fully automated detection and segmentation of lymph nodes on multiparametric-MRI for rectal cancer: a multicenter study” (Ebiomedicine cover article; IF: 11.2): In this paper, our research aimed to address the challenge of accurate and non-invasive evaluation of tumour N staging. We have made a breakthrough in machine recognition of lymph node imaging, pushing the limit to below 3 mm (vs. 5 mm) and achieved intelligent diagnosis of metastasis. Figure 1 shows examples of nodal segmentation displayed on T2WI. A senior editor at The Lancet concluded that this work had high clinical translational value.
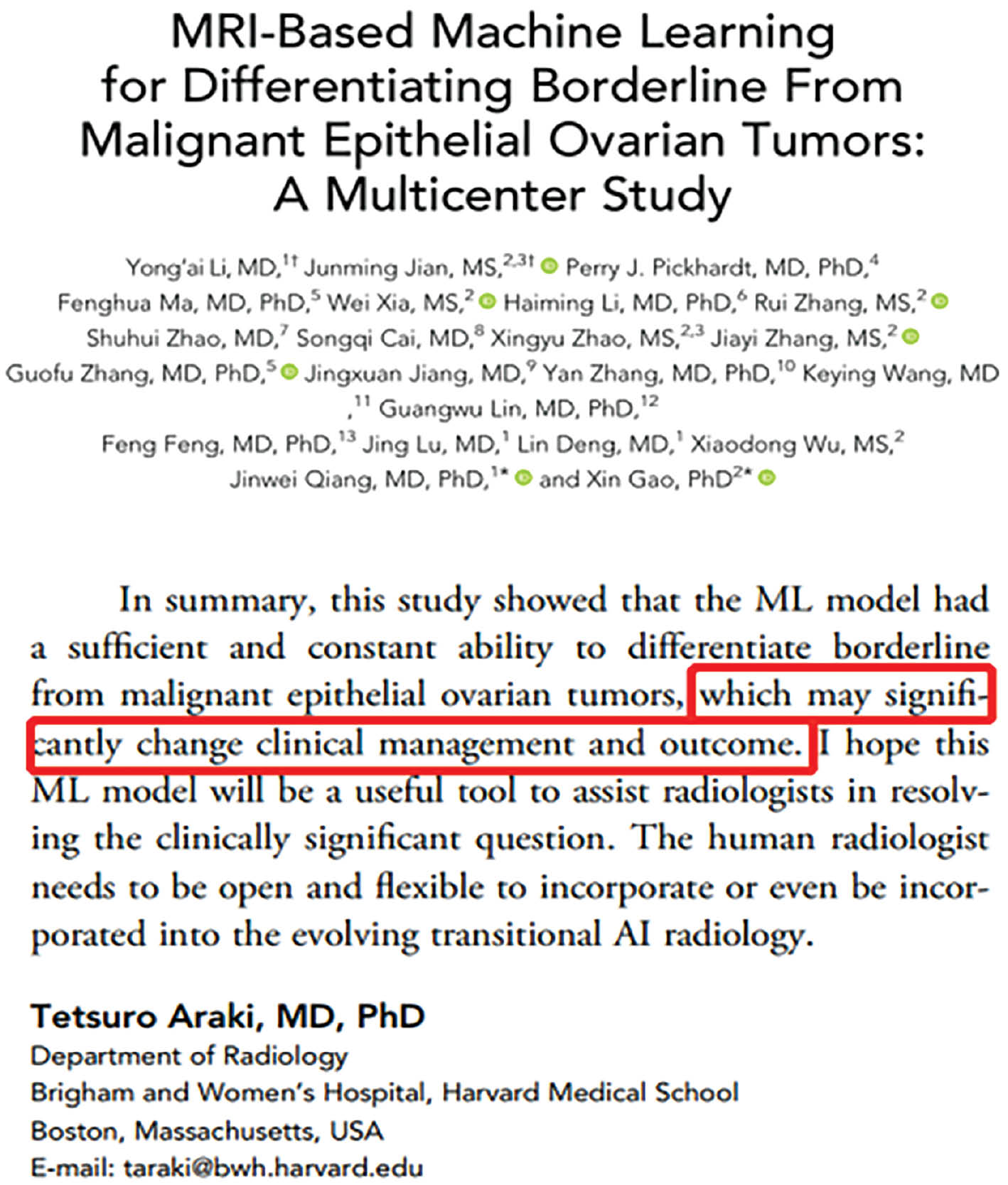
- “MRI-based machine learning for differentiating borderline from malignant epithelial ovarian tumors: a multicenter study”: In this paper, we developed an MRI-based machine learning model for the accurate diagnosis of two subtypes of ovarian tumours. The model was able to achieve an area under the ROC curve of > 0.9, thus outperforming the interpretation of radiologists. Our study was the first attempt to conduct an intelligent non-invasive diagnosis of ovarian cancer, and in so doing, our findings made a significant contribution to an international research gap. Professor Tetsuro Araki from Brigham and Women’s Hospital at Harvard Medical School commented that our work “may significantly change clinical management and outcome” (Figure 2). The relevant achievements of this study have been applied in hospitals, including Jinshan Hospital of Fudan University and Sun Yat-sen University Cancer Center.
Figure 1 Nodal segmentation examples displayed on T2WI. Ground truth results are shown in yellow and segmentation results of the auto-LNDS model are shown in red. The number adjacent to the lymph node is the corresponding DSC.
Figure 2 The comments of Professor Tetsuro Araki from Brigham and Women’s Hospital regarding our work.
Overall, these papers demonstrates the potential of AI and machine learning to improve our understanding of diseases and facilitate the clinical application of precision medicine. I am proud to have contributed to these breakthroughs and look forward to continuing to push the boundaries of this exciting field.
EE: What is the bottleneck in the integration of AI into basic and clinical research? What is the major impasse in the path of your research?
XG: The integration of AI into basic and clinical research is still in the early stages, and there are several bottlenecks that need to be overcome. One of the major challenges is the need for high-quality data, which is crucial for the development and validation of AI models. Obtaining large, high-quality datasets can be time-consuming and resource-intensive. Another challenge is the need for interdisciplinary collaboration because AI research requires expertise in computer science and the application domain, such as medicine or biology. This collaboration can be challenging due to the differences in language and culture between the two fields.
The lack of standardized protocols for the collection and processing of omics data is a barrier to my current research efforts, which can lead to variability in the quality and type of data, and can impact the accuracy and reproducibility of AI models. Addressing this issue requires collaboration across different research groups and the development of standardized protocols for data collection and processing.
EE: Has some of your research been adopted in the clinical setting or currently in use in clinical trials? What preparation is needed for such a “bench-to-bedside” transition? Would you advise other scientists to do the same?
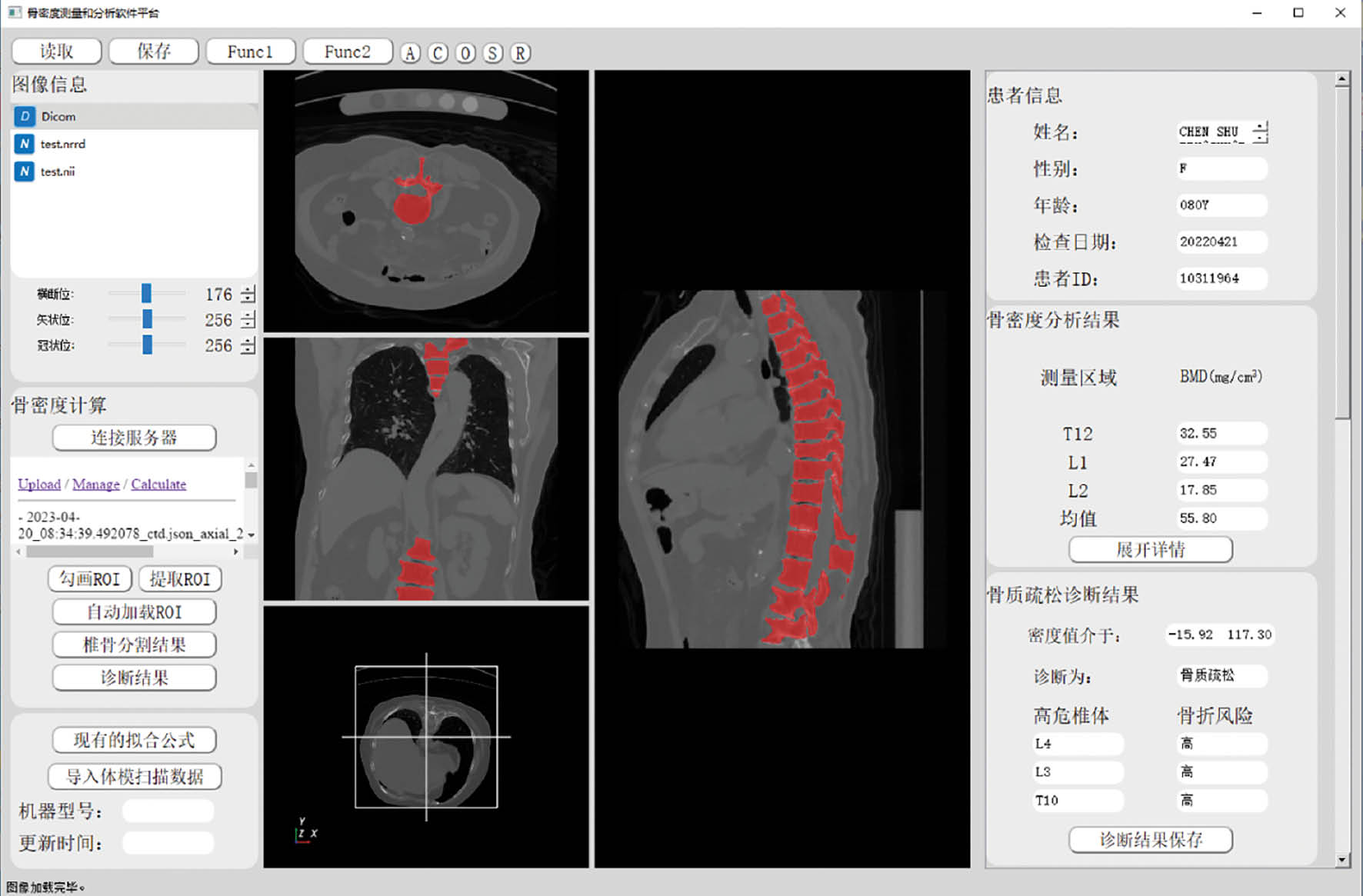
XG: Yes, some of my research has been adopted in the clinical setting and is currently being tested in clinical trials. For example, we developed an intelligent workstation of bone mineral density adopting advanced deep learning algorithms, which can automatically recognize the vertebrae in human CT images and calculate the bone mineral density of vertebral bodies (Figure 3). The preliminary results showed that the accuracy of vertebral recognition exceeded 94%, and agreement between the results from the workstation and manual measurement results of senior physicians reached 90%.
Figure 3 The interface of the intelligent workstation of bone mineral density.
The “bench-to-bedside” transition is a crucial step in ensuring that the benefits of AI research can be realized by patients and clinicians. To achieve this, there needs to be a strong focus on translating research findings into practical clinical tools that can be integrated into existing workflows. One of the key steps in this process is to establish collaborations with clinicians and industry partners to help guide the development of AI tools that meet the needs of patients and healthcare providers. It is also important to consider regulatory requirements and to ensure that any AI-based tools are rigorously evaluated for safety and efficacy.
I would definitely advise other scientists to pursue research that has the potential to be translated into clinical practice. However, it is important to be realistic about the challenges involved in translating research findings into practical applications, and to be willing to engage with clinicians, industry partners, and regulatory agencies to ensure that the transition from “bench-to-bedside” is successful.
EE: What advice do you have for the generation of young scientists today?
XG: My advice for the generation of young scientists today is to be passionate about their research and to pursue their interests with determination and perseverance. I would encourage young scientists to explore new ideas, think creatively, and to be open to collaboration with colleagues from different disciplines. In addition, I believe it is important for young scientists to stay up-to-date with the latest developments in their field and to keep an eye on emerging technologies and methodologies that may be relevant to their research. This requires a commitment to lifelong learning and a willingness to be flexible and adaptable in the face of new challenges and opportunities. Finally, I would urge young scientists to embrace scientific integrity and ethical conduct in their research. It is essential to maintain high standards of rigor and transparency in all aspects of scientific research, and to ensure that findings are reported accurately and honestly. By doing so, we can build trust in the scientific enterprise and make meaningful contributions to our understanding of the world around us.
EE: Do you have any advice for BIO Integration as an emerging journal?
XG: As an emerging journal, my advice for BIO Integration would be to continue to prioritize high-quality research and to build a strong reputation within the scientific community. This can be achieved by maintaining rigorous standards for manuscript review and selection, and by actively seeking out and promoting innovative and impactful research in the field. Additionally, I would encourage BIO Integration to foster a collaborative and inclusive environment that values diversity and promotes open and constructive communication between authors, reviewers, and readers. This can help to build a sense of community and engagement around the journal, and to foster a culture of continuous learning and improvement. Finally, I would suggest that BIO Integration consider embracing emerging technologies and methodologies in scientific publishing, such as open access and preprint servers, to help increase the reach and impact of published research. By staying at the forefront of these trends, BIO Integration can continue to attract high-quality research and establish itself as a leading journal in the field.