Antimalarial Potential of Phytochemical Compounds from Garcinia atroviridis Griff ex. T. Anders Targeting Multiple Proteins of Plasmodium falciparum 3D7: An In Silico Approach
1Department of Biology, Faculty of Science and Technology, Universitas Airlangga, Surabaya, Indonesia
2Postgraduate School, Universitas Airlangga, Surabaya, Indonesia
3Division of Research and Development, Jalan Tengah, Surabaya, Indonesia
4Uttaranchal Institute of Pharmaceutical Sciences, Uttaranchal University, Dehradun, India
5Virtual Research Center for Bioinformatics and Biotechnology, Surabaya, Indonesia
6Doctoral Program in Mathematics and Natural Sciences, Faculty of Science and Technology, Universitas Airlangga, Surabaya, Indonesia
7Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Brawijaya, Malang, Indonesia
8Biology Education Department, Faculty of Teacher Training and Education, Mulawarman University, Samarinda, Indonesia
9Dengue Study Group, Institute of Tropical Diseases, Universitas Airlangga, Surabaya, Indonesia
10Faculty of Medicine, Universitas Wijaya Kusuma Surabaya, Surabaya, Indonesia
11Faculty of Medicine, Universitas Pembangunan Nasional “Veteran” Jawa Timur, Surabaya, Indonesia
12Department of Scientific Research, V. M. Gorbatov Federal Research Center for Food Systems, Moscow, Russian Federation
13Faculty of Biotechnology and Food Engineering, Ural State Agrarian University, Yekaterinburg, Russian Federation
14Department of Scientific Research, Russian State Social University (RSSU), Moscow, Russia
15Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia
16Research Group of Biological Health, Study Program of Biology, Faculty of Health and Science, Universitas Dhyana Pura, Bali, Indonesia
17Department of Biochemistry, Shrimati Indira Gandhi College, Trichy, India
*Correspondence to: Arif Nur Muhammad Ansori, E-mail: ansori.anm@gmail.com; Hery Purnobasuki, E-mail: hery-p@fst.unair.ac.id
Received: September 8 2024; Revised: October 12 2024; Accepted: October 29 2024; Published Online: November 11 2024
Cite this paper:
Aini NS, Ansori ANM, Herdiansyah MA et al. Antimalarial Potential of Phytochemical Compounds from Garcinia atroviridis Griff ex. T. Anders Targeting Multiple Proteins of Plasmodium falciparum 3D7: An In Silico Approach. BIO Integration 2024; 5: 1–12.
DOI: 10.15212/bioi-2024-0075. Available at: https://bio-integration.org/
Download citation
© 2024 The Authors. This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See https://bio-integration.org/copyright-and-permissions/
Abstract
Background: Plasmodium falciparum is a malaria-causing unicellular parasite with strain 3D7 (Pf3D7) being the most lethal. Currently, antimalarial resistance has been reported which necessitates the development of novel antimalarial drugs to combat the spread of malaria. Garcinia atroviridis Griff. ex T. Anders contains phytochemical compounds that are useful for various activities, including targeting Pf3D7 proteins. This study explored novel antimalarial drugs from G. atroviridis against several target proteins of Pf3D7 in silico.
Methods: Phytocompounds from G. atroviridis were selected as ligands. After retrieval from the Protein Data Bank, the protein sequence was screened using BLASTp NCBI. Molecular docking analysis was performed on PyRx to compute binding affinity and identify the chemical interactions involved. The stability of the ligand-protein complex was evaluated using dynamic molecular approaches.
Results: Our findings showed that quercetin has a high binding affinity with apicoplast DNA polymerase (−8.3 kcal/mol), glutamyl-tRNA synthetase (−7.5 kcal/mol), and plasmepsin X (−7.8 kcal/mol). Kaempferol had a high binding affinity for the cytochrome c2 domain-swapped dimer (−8.4 kcal/mol).
Conclusion: Collectively, quercetin and kaempferol are potential antimalarial candidates which warrant further investigation using in vitro and in vivo designs.
Keywords
Bioinformatics, Drug Discovery, Malaria, Medicine, Molecular Docking
Introduction
Malaria is one of the deadliest infectious diseases with global cases spanning different age groups [1]. According to data from the World Health Organization (WHO) in 2020, malaria is endemic in >50 countries, including Indonesia [2, 3]. Malaria is caused by the protozoan parasite, Plasmodium, which is transmitted to humans from the bites of female Anopheles mosquitoes [4, 5]. P. falciparum is a parasite responsible for major malaria cases worldwide, particularly in sub-Saharan Africa and Asia [6, 7]. P. falciparum is a unicellular protozoan belonging to the Plasmodiae family and Apicomplexa phylum [8]. Among thousands of identified strains, P. falciparum 3D7 (Pf3D7) is considered the most lethal [9]. Management of malaria is challenged by the presence of antimalarial drug resistance, such as chloroquine, artemisinin, and sulfadoxine-pyrimethamine, which has been reported in developed countries [10, 11].
To address this challenge, research has been conducted to explore phytocompounds that possess biological activities and drug-likeness [12]. Artemisinin and quinine are examples of such discoveries [13]. G. atroviridis Griff. ex. T. Anders has been specifically reported for its use in ethnomedicine in South and Southeast Asian countries [14, 15]. Previous studies have reported that the extract of G. atroviridis has antioxidant, antimicrobial, and anticancer activities [16–18]. The leaf extract of G. atroviridis has been reported to act as an effective inhibitor of P. berghei parasitemia in a mouse model [19]. However, no further research has been conducted to investigate the potential of phytocompounds from G. atroviridis as antimalarial agents using in vivo, in vitro, or even in silico approaches [20].
In silico is considered the method of choice in the drug discovery process because of its efficiency in terms of economic cost and time consumption. Moreover, molecular docking provides insight in the predicted orientation and position of drug candidates as potential substrates for target molecules [8, 21, 22]. The target proteins include PfapPOL (PDB ID: 7SXQ), Pf pyruvate kinase complex (PDB ID: 7Z4M), Pf actin 1 filament (PDB ID: 6TU4), Pf aspartate transcabamoylase (PDB ID: 7ZCZ), PfERS (PDB ID: 7WAJ), Pfcyt c2 DSD (PDB ID: 7TXE), and PfPMX (PDB ID: 7RY7). All of the aforementioned proteins are Pf3D7 receptors that can debilitate the P. falciparum 3D7 body. These proteins are crucial receptors in the Pf3D7 strain that determine the survival and virulence of the organism and serve as a preliminary study to investigate the potential of G. atroviridis-based compounds as antimalarial drug candidates.
Methods
Ligand collection
The phytochemical compounds from G. atroviridis, which are based on previous studies conducted by Shahid et al. (2022), were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) [15]. The identification and selection of phytochemical compounds from G. atroviridis were performed using ChemDraw to obtain the 2D structure and Chem3D to visualize the 3D structure. The 3D structure was retrieved from. sdf format. The compound identifier (CID) and canonical SMILES of each compound were collected from the PubChem database. Ligand minimization was conducted using Open Babel in PyRx software (version 0.9;The Scripps Research Institute, La Jolla, CA, USA) [23].
Antimalarial probability prediction
Canonical SMILES of each compound was pasted on the PASS Online webserver to determine antimalarial potency. Potential activity (Pa) > Potential inactivity (Pi), and Pa >0.3 were set as the standard. Compounds with these values for one or two categories in antiprotozoal and antimalarial activities are thought to operate properly in the human body [24].
Drug absorption parameter analysis
Selective antimalarial potential phytochemical compounds from G. atroviridis were compared of the drug similarity using SwissADME (http://www.swissadme.ch/index.php). Lipinski rules were utilized to identify the drug absorption parameter in this analysis with categories comprising hydrogen bond donor (nOHNH) ≤5, hydrogen bond acceptor (nON) ≤10, coefficient partition in water-lipid (miLogP) ≤5, and relative molecular mass (BM) ≤500 [25, 26].
Target protein collection
Target proteins were collected from RCSB PDV (https://rcsb.org/) in the P. falciparum 3D7 (Pf3D7) category. Recent publications from 2022–2023 were collected. The protein was then identified by comparative sequence analysis using the BLASTp tool in NCBI [27]. A threshold value ≤35% was established for sequence identity [28]. A low significance of similarity demonstrates a low homology with human proteins [8, 29].
Molecular docking, validation protocol, and complex visualization
Molecular docking was performed to screen the interaction between the ligand and the target protein via specific docking [22]. The process was performed using PyRx software (version 0.9; The Scripps Research Institute, La Jolla, CA, USA) [23]. Moreover, the binding affinity score and position of the most negative phytochemical ligands were collected and compared [30]. To validate the stability of the molecular docking method, molecular dynamics were assessed with CABS-flex 2.0 (https://biocomp.chem.uw.edu.pl/CABSflex2/index) [31]. The simulation parameters included protein rigidity, restraints, C-alpha restraint weight, number of cycles, side-chain restraints, temperature range, trajectory, and RNG seed [32, 33]. Next, the selected ligands with predicted target proteins were displayed using Biovia Discovery Studio software (Vélizy-Villacoublay, France). The selected ligands were visualized in 2D and 3D to determine the details of the binding site interaction between the ligand and the target protein [8].
The crystallographic structure conformer of each target protein with the natural ligand was docked to the receptor using AutoDock Tools (PyRx software, version 0.9; The Scripps Research Institute, La Jolla, CA, USA) by setting a grid box scale. If the root mean square deviation (RMSD) value was >2 Å, the procedure was invalid. Therefore, the grid boxes (X, Y, and Z) and spacing values (center X, Y, and Z) were manually adjusted until a RMSD <2 Å was obtained. The validation molecular docking process needs to determine the grid box to understand the interaction of the ligand and protein as the active site of the protein. The center of the grid box was determined based on the center of mass of the naturally occurring ligand. The dimensions of the grid box were based on the size of the ligand and binding site (Table 1) [34, 35].
Table 1 Grid Box Scale of Molecular Docking
| Target protein | Ref | Grid box scale | |||||
|---|---|---|---|---|---|---|---|
| Center (Å) | Dimension (Å) | ||||||
| X | Y | Z | X | Y | Z | ||
| 7SXQ | [36] | −50.263 | 3.428 | 15.813 | 117.116 | 91.309 | 118.546 |
| 7WAJ | [37] | −27.384 | −9.320 | 20.247 | 71.059 | 85.187 | 101.557 |
| 7TXE | [38] | −13.662 | −4.523 | 14.643 | 72.095 | 67.269 | 42.993 |
| 7RY7 | [39] | 68.445 | 5.069 | 39.239 | 87.576 | 62.283 | 77.647 |
Results
G. atroviridis contains 35 bioactive phytocompounds derived from different parts of the plant, as suggested by a previous comprehensive critical review. Some parts of G. atroviridis have been reported as a source of bioactive phytocompounds, including fruits, stem bark, and leaves [15]. The structure of each compound was retrieved from the PubChem database, where only 24 of the structures had a CID and canonical SMILES notation (Table 2).
Table 2 Phytochemical Compounds of G. atroviridis from Previous Research [15]
| Compound | Sources | PubChem CID | References |
|---|---|---|---|
| Citric acid | Fruit | 311 | [73, 74] |
| Malic acid | Fruit | 525 | [73, 75] |
| Succinic acid | Fruit | 1110 | [73, 76] |
| Tartaric acid | Fruit | 875 | [73] |
| Hydroxycitric acid | Fruit | 123908 | [77–79] |
| Pentadecanoic acid | Fruit | 13849 | [73, 80–83] |
| Nonadecanoic acid | Fruit | 12591 | [73] |
| Dodecanoic acid | Fruit | 3893 | [73] |
| 14-cis-docosenoic acid | Fruit | Unknown | [84] |
| 1,1″-dibutyl methyl hydroxycitrate | Fruit | Unknown | [85] |
| 2-(butoxycarbonylmethyl)-3-butoxycarbonyl-2-hydroxy-3-propanolide | Fruit | Unknown | [85] |
| Atroviridin | Stem bark | 11267348 | [86] |
| Benzoquinone atroviridine | Root | Unknown | [87, 88] |
| Atrovirisidone | Root | 10342405 | [88] |
| Atrovirisidone B | Root | Unknown | [88] |
| Garcineflovanol A | Stem bark | Unknown | [89] |
| Garcineflovanone A | Stem bark | Unknown | [89] |
| Naringenin | Root | Unknown | [90] |
| 3,8″-binaringenin | Root | Unknown | [90] |
| Morelloflavone | Root | 5464454 | [84] |
| Fukugiside | Root | 73157060 | [84] |
| Kaempferol | Stem bark | 5280863 | [91] |
| Quercetin | Stem bark | 5280343 | [91] |
| Garcinol | Fruit | 5281560 | [91] |
| Isogarcinol | Fruit | 11135781 | [91] |
| α-humulene | Fruit | 5281520 | [92] |
| (−)-β-caryophyllene | Fruit | 1742210 | [92] |
| Cambroginol | Fruit | Unknown | [73] |
| 4-methylhydroatrovirinone | Root | 101249096 | [84] |
| Garcinexanthone G | Stem bark | Unknown | [91] |
| Gentisein | Stem bark | 5281635 | [91] |
| Stigmasterol | 5280794 | [91] | |
| Stigmasta-5,22-dien-3-O-β-glucopyranoside | Stem bark | Unknown | [91] |
| 3β-acetoxy-11α,12α-epoxyoleanan-28,13β-olide | Stem bark | Unknown | [91] |
| 2,6-dimethoxy-p-benzoquinone | Stem bark | 68262 | [91] |
Selected phytochemical compounds with known CIDs were analyzed to identify the anti-protozoal and anti-Plasmodium probabilities using PASS online. From the probability screening using PASS online, there were five compounds that matched the criteria as antimalarials: fukugisisde (CID 73157060); kaempferol (CID 5280863); quercetin (CID 5280343); (−)-β-caryophyllene (CID 1742210); and gentisein (CID 5281635). All selected compounds are shown in Table 3. Kaempferol and quercetin had the highest scores for both anti-protozoal and anti-Plasmodium agents compared to fukugiside, (−)-β-caryophyllene, and gentisein, which only fulfilled the criteria for anti-protozoal probability. Based on the Lipinski rules, fukugiside received three violations and was discarded from the subsequent docking analysis (Table 3). Drug absorption analysis was performed to determine the possibility of drug absorption in the body (Table 4).
Table 3 Antiprotozoal and Anti-Plasmodium Probabilities of Phytochemical Compounds from G. atroviridis Collected from PASS Online
| Compounds | Antiprotozoal | Anti-plasmodium | ||
|---|---|---|---|---|
| Pa | Pi | Pa | Pi | |
| Ascorbic acid | – | – | – | – |
| Citric acid | – | – | – | – |
| Malic acid | – | – | – | – |
| Succinic acid | – | – | 0.175 | 0.084 |
| Tartaric acid | 0.025 | 0.007 | – | – |
| Hydroxycitric acid | – | – | – | – |
| Pentadecanoic acid | 0.019 | 0.007 | 0.181 | 0.076 |
| Nonadecanoic acid | 0.019 | 0.007 | 0.181 | 0.076 |
| Dodecanoic acid | 0.019 | 0.007 | 0.181 | 0.076 |
| Atroviridin | 0.211 | 0.096 | 0.192 | 0.065 |
| Atrovirisidone | 0.248 | 0.072 | 0.224 | 0.039 |
| Morelloflavone | 0.194 | 0.111 | 0.176 | 0.083 |
| Fukugiside | 0.342 | 0.033 | 0.298 | 0.014 |
| Kaempferol | 0.461 | 0.013 | 0.345 | 0.009 |
| Quercetin | 0.446 | 0.014 | 0.365 | 0.008 |
| Garcinol | 0.298 | 0.047 | – | – |
| Isogarcinol | – | – | – | – |
| α-humulene | – | – | – | – |
| (−)-β-caryophyllene | 0.578 | 0.006 | 0.231 | 0.034 |
| 4-methylhydroatrovirinone | 0.205 | 0.100 | 0.193 | 0.064 |
| Genistein | 0.335 | 0.034 | 0.262 | 0.022 |
(-) No probability of antiprotozoal or anti-plasmodium candidate.
Table 4 Drug Absorption Analysis of Selected Phytochemical Compounds from G. atroviridis
| Compounds | nOHNH (≤5) | nON (≤10) | miLogP (≤5) | BM (≤500 g/mol) | Violation |
|---|---|---|---|---|---|
| Fukugiside | 16 | 10 | 1.62 | 718.61 | 3 |
| Kaempferol | 6 | 4 | 2.17 | 286.24 | 0 |
| Quercetin | 7 | 5 | 1.68 | 302.24 | 0 |
| (−)-β-caryophyllene | 1 | 0 | 4.14 | 220.35 | 0 |
| Genistein | 5 | 3 | 2.27 | 244.20 | 0 |
In contrast, 514 target protein structures retrieved from Pf3D7 were collected from RCSB PDB. Seven selected proteins with native ligands were filtered from 2022–2023 publications and compared to human homology proteins using BLASTp NCBI (Table 5).
Table 5 Target Protein Screening
| Protein target (PDB ID) | Resolution (Å) | Homology protein (accession number) | Native ligand similarity (%) |
|---|---|---|---|
| Pf apicoplast DNA polymerase (7SXQ) | 2.50 | 4XVL_A | 23.67 |
| Pf pyruvate kinase complex (7Z4M) | 1.90 | 5SC8_A | 47.77 |
| Pf actin 1 filament (6TU4) | 2.60 | 7P1H_B | 83.06 |
| Pf aspartate transcabamoylase (7ZCZ) | 2.45 | 5G1N_A | 36.31 |
| Pf glutamyl-tRNA synthetase (PfERS) (7WAJ) | 2.25 | 4YE8_A | 31.11 |
| Pf cyt c2 DSD (7TXE) | 2.30 | 5EXO_A | 32.14 |
| Pf plasmepsin X (PMX) (7RY7) | 2.10 | 3O9L_A | 32.72 |
Four proteins were obtained with similarities <35%, including apicoplast DNA polymerase [apPOL] (PDB ID 7SXQ), glutamyl-tRNA synthetase [ERS] (PDB ID 7WAJ), cytochrome c2 domain-swapped dimer [cyt c2 DSD] (PDB ID 7TXE), and plasmepsin X [PMX] (PDB ID 7RY7). Filtered proteins were further utilized in docking analysis to identify ligand-target protein interactions (Table 5) [36–39].
Molecular docking analysis was used to determine the binding affinity and chemical interactions of amino acids between the ligand and target protein. The lowest binding affinity was quercetin. Quercetin had the lowest binding affinity against apPOL, ERS, and PMX (−8.3, −7.5, and −7.8 kcal/mol, respectively). Kaempferol had the lowest result compared to other selected compounds targeting cyt c2 DSD (−8.4 kcal/mol; Table 6).
Table 6 Chemical Interaction between Ligand and Target Protein Complex
| Ligand-protein complex | Binding affinity (kcal/mol) | Interaction | Amino acids |
|---|---|---|---|
| Quercetin−apPOL | −8.3 | HI | Lys29(A), Ile76(A), Tyr105(A) |
| PHI | Lys29(A), Lys74(A), Lys77(A), Tyr78(A), Glu103(A) | ||
| vdw | Lys27(A), Leu28(A), Ile72(A), Cys79(A), Asn104(A) | ||
| UF | Tyr105(A) | ||
| Quercetin−ERS | −7.5 | HI | Leu576(A), Arg577(A) |
| PHI | Asn371(A), Leu580(A), Lys785(A), Asp797(A), Asp797(A) | ||
| vdw | Thr581(A), Lys582(A), Val782(A), Ser783(A), Ile795(A), Ile808(A) | ||
| Quercetin-Cyt c2 DSD | −8.2 | HI | Pro60(A) |
| PHI | Asn52(A), Arg68(A) | ||
| Vdw | Leu97(A), Met101(A), Val65(A), His42(A), Ala70(A), Gly71(A), Thr51(A), Lys74(A), Trp58(B), Leu132(B) | ||
| Quercetin-PMX | −7.8 | HI | Asp112(A), Ile358(A) |
| PHI | Asp245(A), Gln247(A), Tyr462(A) | ||
| vdw | Asn72(A), Asp73(A), His74(A), Thr110(A), Leu111(A), His242(A), Asp356(A), Tyr357(A), Ser359(A) | ||
| UF | Pro71(A) | ||
| Kaempferol-apPOL | −8.1 | HI | Tyr105(A) |
| PHI | Lys29(A), Ile72(A) | ||
| vdw | Tyr78(A), Glu103(A), Val102(A), Asn104(A), Asp75(A), Lys74(A), Lys27(A), Leu28(A) | ||
| UF | Lys77(A) | ||
| Kaempferol-ERS | −7.3 | PHI | Glu669(A), Ser628(A), Thr769(A) |
| vdw | Asp625(A), Leu670(A), Glu671(A), Arg790(A), Asp672(A), His723(A) | ||
| Kaempferol-Cyt c2 DSD | −8.4 | HI | Pro60(A), Pro60(A), Leu62(A), Trp92(A), Trp92(A), Tyr100(A) |
| PHI | Arg68(A), Arg68(A) | ||
| UF | Asn52(A) | ||
| Kaempferol-PMX | −7.4 | PHI | Pro71(A), His242(A), Asp73(A), Tyr357(A) |
| vdw | Ile358A), Asp112(A), Leu111(A), Lys76(A), Tyr462(A), His74(A), Gln247(A), Asn72(A), Arg244(A), Ser359(A), Thr110(A) | ||
| (−)-β-caryophyllene-apPOL | −6.8 | HI | Lys244(A), Leu557(A), Ile241(A) |
| vdw | Asn603(A), Leu557(A), Tyr607(A), Ile237(A) | ||
| (−)-β-caryophyllene-ERS | −6.1 | HI | Arg509(A), Leu508(A), Asn334(A), Thr510(A), His327(A), Asp350(A), Pro317(A), Glu318(A) |
| vdw | Tyr491(A), Pro316(A), Phe315(A), Phe537(A), Ala330(A) | ||
| (−)-β-caryophyllene-Cyt c2 DSD | −6.5 | HI | Lys74(A), Phe79(A) |
| vdw | Thr87(A), Ser77(A), Pro78(A), Ser73(A), Thr69(A), Phe55(A) | ||
| (−)-β-caryophyllene-PMX | −6.6 | HI | Ile99(A), Ile316(A), His242(A), Val340(A), Phe360(A) |
| vdw | Lys241(A), Trp273(A) | ||
| Gentisein-apPOL | −7.9 | PHI | Thr85(A), Asn139(A), Gln138(A), Ile83(A), Asp143(A). Asn82(A) |
| vdw | Gln195(A), Trp199(A). Thr86(A), Gln84(A), Phe142(A), Leu178(A) | ||
| Gentisein-ERS | −6.7 | vdw | Ser628(A), Asp625(A), Thr769(A), Arg790(A). Asp672(A) |
| Gentisein-Cyt c2 DSD | −7.8 | HI | Lys39(A) |
| PHI | Ser77(B) | ||
| vdw | Ser57(A), Gln40(A), Tyr149(B), Thr(56), Gly75(B), Lys74(B), Asn76(B) | ||
| Gentisein-PMX | −7.5 | HI | Leu84(A) |
| PHI | Gln527(A), Val89(A), Asn475(A) | ||
| vdw | Ser467(A), Lys90(A), Tyr91(A), Met470(A), Leu474(A) |
HI: hydropobic interaction; PHI: polar H interaction; vdw: van der Walls interaction; UF: unfavorable acceptor.
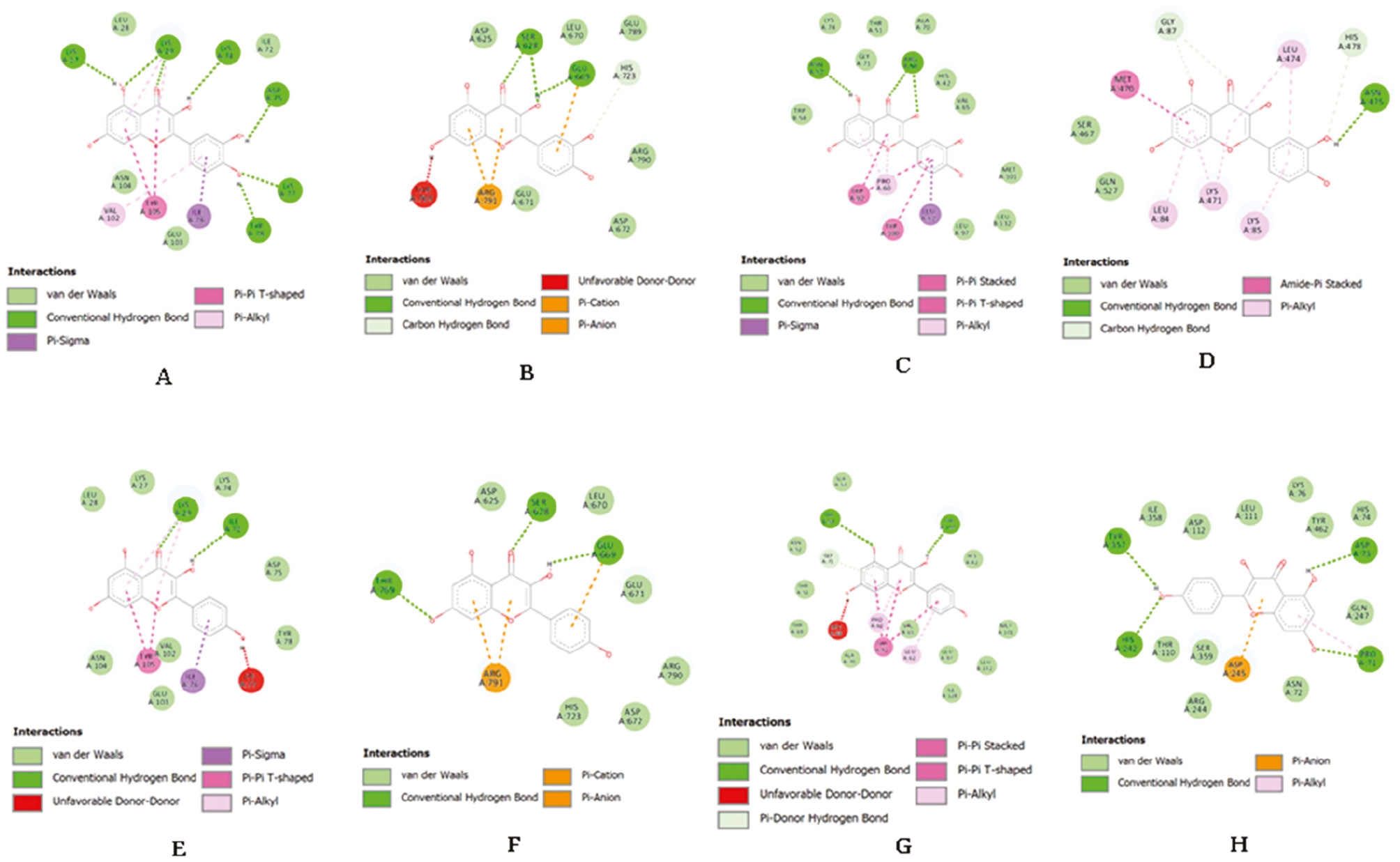
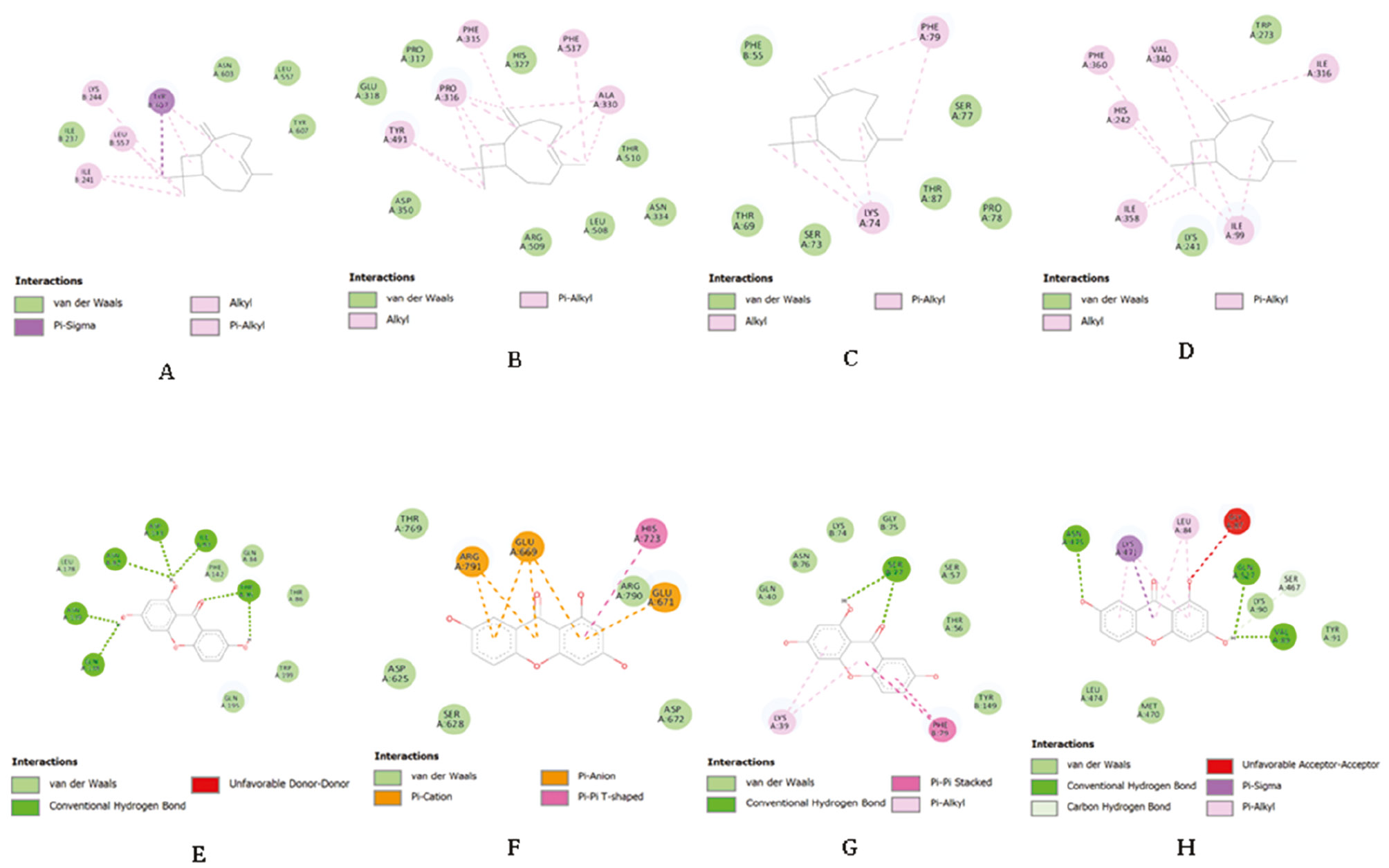
Visualization of ligand-target protein interaction is shown with red stain for the target protein and zoomed in to display the ligand interaction. Chemical interactions between ligands binding to the A domain of each protein are shown. Hydrogen bonds, hydrophobic, and van der Waals (vdw) interactions formed between compounds and target proteins. Conventional hydrogen bonds, pi-donor hydrogen bonds, pi-sigma, pi-pi T-shaped, pi-alkyl, pi-pi stacked, and amide-pi stacked consist of various hydrogen bonds and hydrophobic interactions were involved in the four selected target proteins (Figures 1 and Figures 2). However, only the quercetin and ERS complex interaction did not result in an unfavorable acceptor-acceptor bump (Table 6).
Figure 1 Chemical interaction between receptor and compounds (A: quercetin−apPOL; B: quercetin−ERS; C: quercetin-cyt c2 DSD; D: quercetin-PMX; E: kaempferol-apPOL; F: kaempferol-ERS; G: kaempferol-cyt c2 DSD; H: kaempferol-PMX.
Figure 2 Chemical interaction between receptor and compounds [I: (−)-β-caryophyllene-apPOL; J: (−)-β-caryophyllene-ERS; K: (−)-β-caryophyllene-cyt c2 DSD; L: (−)-β-caryophyllene-PMX; M: gentisein-apPOL; N: gentisein-ERS; O: gentisein-cyt c2 DSD; P: gentisein-PMX].
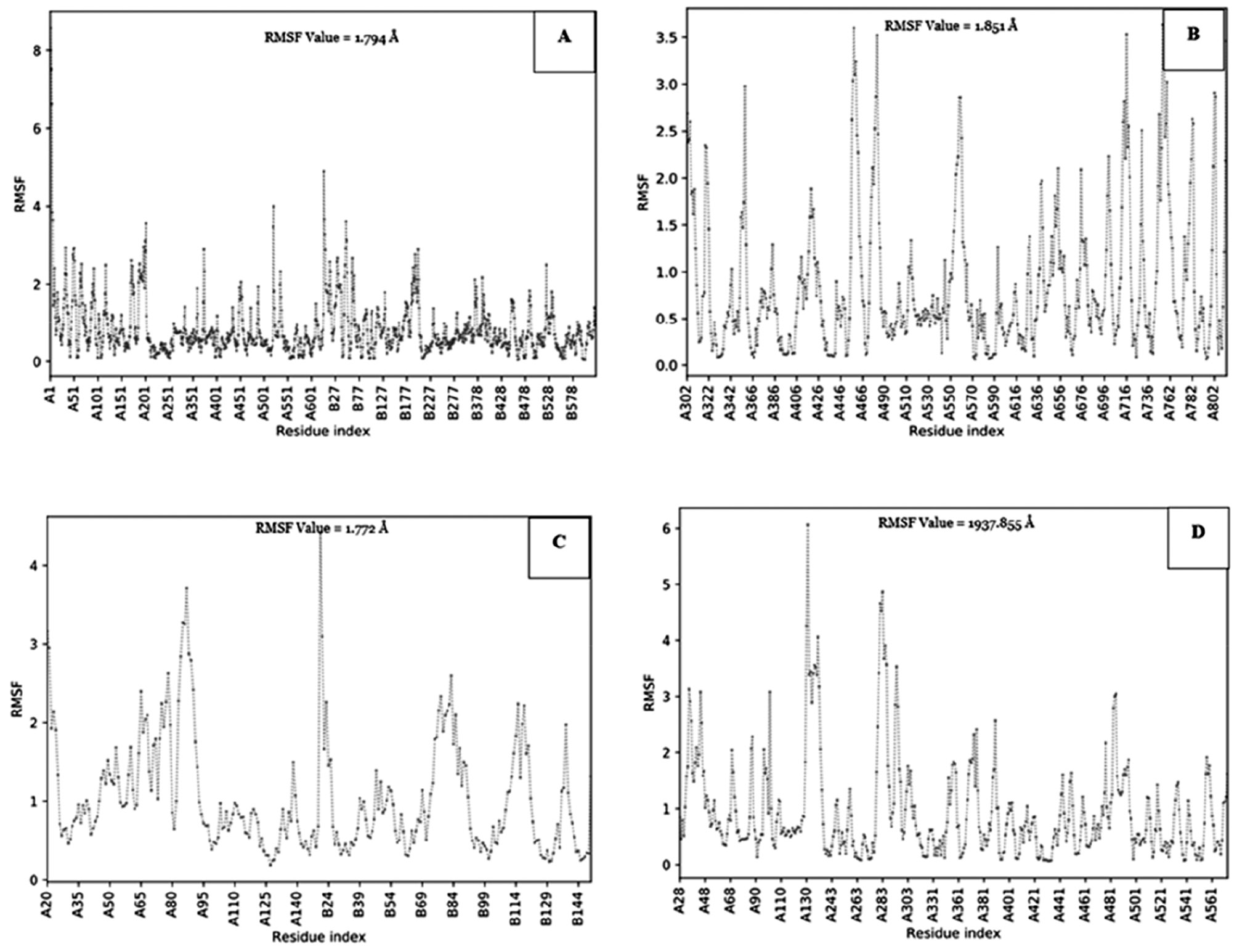
Validation docking was also available in this study to determine the stability of the receptor when applied with the compound candidate. Molecular dynamic analysis results indicated that the interaction hotspot total root mean square fluctuation (RMSF) value was different for each of the receptors. The most stable receptor was 7TXE, with stable fluctuations between the ligand and protein atoms. Receptor 7TXE had an RMSF value <3 Å (Figure 3C). The receptor with the code, 7RY7, was the most unstable receptor against the compound because the fluctuations formed from the ligand and protein atoms had an RMSF value >3 Å (Figure 3D). Thus, all molecular dynamic validations are shown in Figure 3.
Figure 3 Molecular docking validation of the receptor using the root mean square fluctuation (RMSF) value through CABS-flex online tools (A: 7SXQ; B: 7WAJ; C: 7TXE; D: 7RY7).
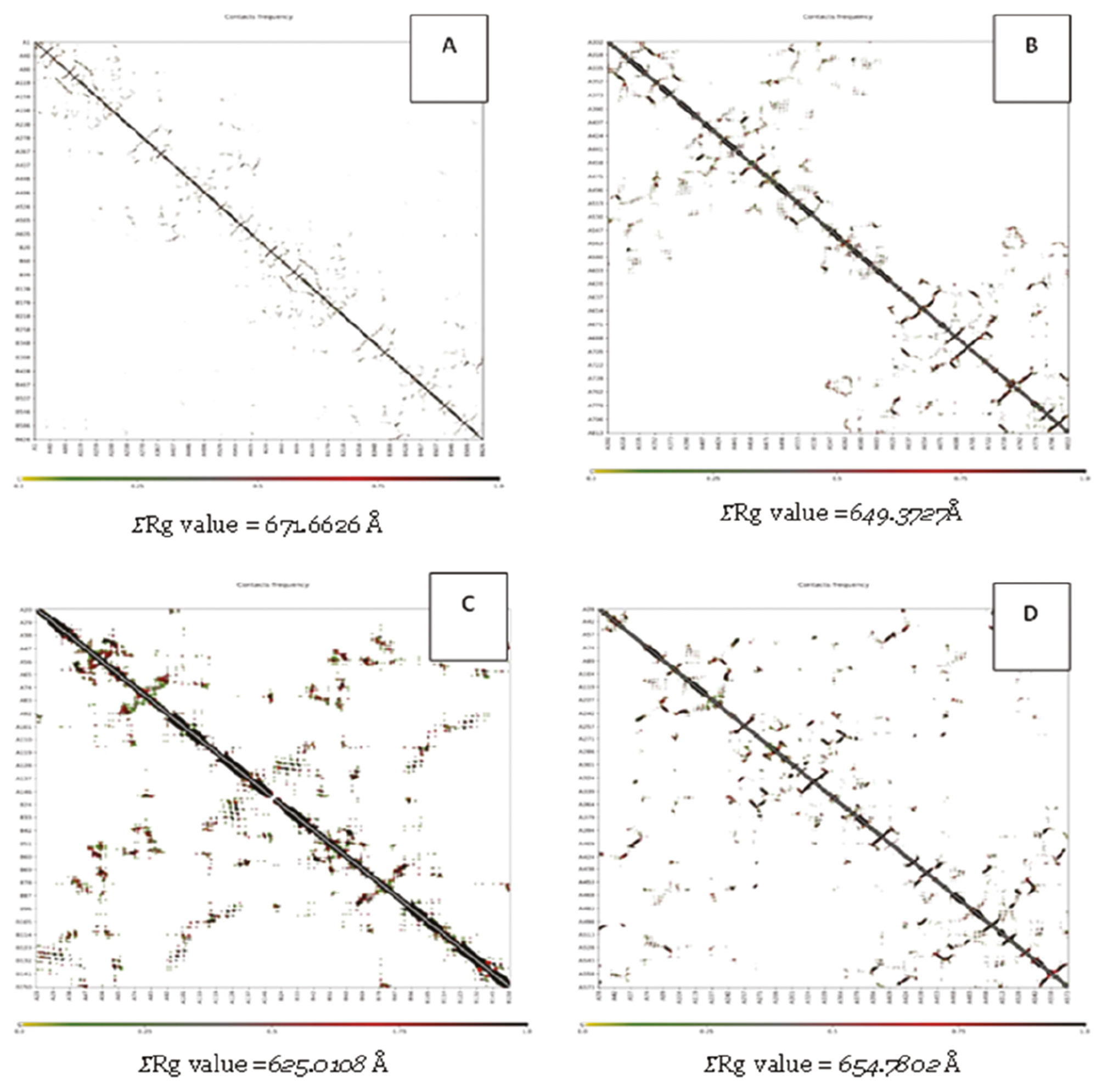
Another docking simulation validation revealed the radius of gyration (Rg) for each receptor (Figure 4). As shown in Figure 4, the Rg of the complex receptor fluctuated between 0.100 and 1.000 Å. Rg showed little conformational change throughout the docking simulation [40]. A lower Rg value indicates that the system has higher compactness and vice versa [41]. The data in Figure 4 show that that 7TXE complex was the most stable, with a low Rg value. The 7SXQ complex had the most unstable interaction with the highest Rg value.
Figure 4 Molecular docking validation of the receptor using the radius of gyration (Rg) value through CABS-flex online tools (A: 7SXQ; B: 7WAJ; C: 7TXE; D: 7RY7).
Discussion
G. atroviridis has numerous benefits for various diseases [15]. The methanol extract from this plant showed better antioxidant activity than the aqueous extract. Among the various parts, methanol extracts from the stem had the highest total phenolic and flavonoid content as well as the strongest antioxidant extract based on 1,1-diphenyl-2-picrylhydrazyl (DPPH) and 2,2-azinobis 3-ethylbenzothiazoline 6-sulfonate (ABTS) scavenging assays, respectively. This finding demonstrated a significant correlation between phytochemical constituents that are responsible for radical scavenging and antioxidant effects [16]. In contrast, the antimicrobial activities of G. atroviridis phenol extract had a 6.67%–42.86% inhibition for various fungi that resembled the antioxidant properties with moderate inhibition to selective fungi and cell viability targeting human skin fibroblast (HSF) cells [17]. Two new ester derivatives of garcinia acid showed anti-tumor promoting activity against Epstein-Barr virus early antigen and non-cytotoxic characteristics towards several tested cell cultures [18]. In the current study the PubChem webserver collected restricted databases that facilitated detection of antimalarial probability for 23 compounds and showed good criteria for 5 compounds. Furthermore, the drug absorption analysis revealed that only four phytochemical compounds fulfilled these criteria.
In contrast, one study targeting Plasmodium parasitemia utilized G. atroviridis. However, the research was conducted to treat P. berghei parasitemia in mice via in vivo research and no records of specific phytochemical compounds suppressing parasitemia have been published [19]. There are 514 Pf3D7 protein structures deposited in the RCSB PDB. Several proteins have been identified, including plasmepsins, proteases, peptidases, and purine nucleosides [10]. A recently published protein was required to determine the similarity with homologous proteins from humans to select appropriate targets for antimalarial drugs. Following publication and homology selection, selected proteins were docked to select phytochemical compounds from G. atroviridis as ligands [8].
Apicoplast is non-photosynthetic plastid that has evolved from chloroplasts. This organelle arose through a secondary endosymbiotic event in with red algae [36]. The apicoplast participates in metabolic processes, such as fatty acid, heme, and isoprenoid biosynthesis, as well as Fe-S maturation [42, 43]. Furthermore, DNA polymerase targeting apPOL has a role in genome replication and repair. BLASTp analysis revealed the lowest similarity to orthologs in mammals. Therefore, this protein is a promising drug target for malaria prevention and treatment [36]. Based on molecular docking analysis, domain A of this protein is critical for replication inhibition. Quercetin interacts against apPOL in various ways, including hydrogen bonding and hydrophobic and vdw interactions (Table 6). Hydrogen bonds in this complex involved seven interactions, including conventional and pi-donor hydrogen bonds. The hydrogen bond exhibits the strongest chemical interaction [44]. Quercetin was predicted to be the most effective inhibitor of apPOL activity in this study.
Aminoacyl-tRNA synthetases (aaRSs) are vital enzymes in protein translation that charge tRNA for protein synthesis [45]. Most aaRSs do not require tRNA to produce amino acid and adenosine monophosphate (aa-AMP) complexes in the translation process. However, there are some exceptions at this stage, such as glutamyl- (GluRS), glutaminyl- (GlnRS), and arginyl-tRNA (ArgRS) synthetases [46, 47]. ATP is triggered by tRNA binding in these enzymes and forms an adenylate intermediate complex [48, 49]. There are several functional structures (pocket regions, cavities, and tunnels) that connect protein surfaces with buried active or binding sites in protein conformers. This structure is essential for the biological activity of most proteins [50]. Cytoplasmic PfERS has significant evolutionary divergence through the process of L-Glu production and has been studied as an antimalarial agent for decades [51]. Molecular docking results showed different interactions, such as hydrogen bonds and hydrophobic and vdw interactions. However, quercetin did not exhibit unfavorable bonds that provided a less satisfactory binding affinity throughout the molecular docking [52]. Compared to other phytochemical compounds, quercetin had the lowest binding affinity for inhibiting Pf3D7 cytoplasmic L-Glu biosynthesis.
P. falciparum attacks erythrocytes and degrades hemoglobin into crystalline hemozoin within the acidic parasite digestive vacuoles [53]. Heme from hemoglobin is an essential metabolic cofactor for P. falciparum. Parasites preserve a mitochondrial electron transport chain (ETC), including cyt c, as a mobile electron carrier between complexes III and I [31]. Complex III binds the reduced ubiquinol and transports electrons via the Q cycle reaction. The ETC function is critical for parasite viability, particularly ATP synthesis [54]. According to the docking analysis, hydrophobic interactions predominate in the kaempferol chemical interaction with cyt c2 DSD. Pi-sigma, pi-pi stacked, pi-pi T-shaped, and pi-alkyl interactions are hydrophobic interactions. Hydrophobic interactions are known to dominate protein stability in proteins with 36–534 residues, accounting for approximately 60±4% of all interaction [55]. Thus, the interaction between kaempferol and cyt c2 DSD is regarded as the most potent inhibitor of hemoglobin degradation and ATP synthesis compared to other phytochemical compounds from G. atroviridis.
Pepsin-like aspartic proteases influence nutrient uptake, immune evasion, invasion, and egress, which are important processes for successful infection of the host cell [39]. P. falciparum, the most lethal Plasmodium species, expresses 10 pepsin-like aspartic proteases in plasmepsin (PfPM) [9]. Among the pepsin-like aspartic proteases, PfPMIX and PfPMX are involved in parasite invasion and egress [56, 57]. PMX is located in exonemes or secretory vesicles and is expressed in the schizonts, merozoites, gametocytes, and liver infections [39, 58]. Like previous results of selected ligands targeting essential target proteins of Pf3D7, quercetin chemical interactions include hydrogen bonds, hydrophobic interactions, vdw interactions, and an unfavorable bump. Vdw interactions are abundant, although the consecutive functions of vdw are weak. However, the close atomic and molecular distances encourage vdw interactions in the compactness and folding of secondary structured proteins [59]. Moreover, quercetin may have potential as a PMX preventive agent at various stages of parasitemia.
Previous studies have revealed numerous agents with selective targets against several proteins in P. falciparum. Various antibiotics, such as clindamycin and tetracycline, inhibit parasite differentiation into merozoites [60, 61]. The antibiotics may impede aminoacyl-tRNA binding to mRNA ribosomes and inhibit the protein synthetase pathway. Quinoline derivatives act via heme detoxification and the cytochrome BCI complex. However, several antimalarial drugs, such as artimisinin and primaquine, have unclear mechanisms [11, 13, 62]. In addition, resistance to synthetic drugs has been reported since 1957 in Southeast Asia and sub-Saharan Africa [63, 64].
Furthermore, the phytochemical compounds of G. atroviridis have been computationally approved for the antimalarial potential. Quercetin has a wide range of targets against proteins in various organelles, such as apicoplast (apPOS), cytoplasm (ERS), and vacuole (PMX). Kaempferol has been shown to prevent cyt c2 DSD activities of Pf3D7. Both quercetin and kaempferol are plant-derived aglycones (flavonol) from flavonoid glycosides that are synthesized by different enzymes. Quercetin and kaempferol also demonstrated broad spectrum functions, such as antioxidant, antimicrobial, antitumor, cardiovascular protection, and anti-inflammatory activities [65–67]. The pharmacokinetics and toxicology assessments of quercetic and kaempferol have been categorized as safe in several dosage amounts [65, 68]. Further exploration should be conducted regarding the potential of these compounds to enrich the database of phytochemical and bioinformatic medicinal function [31].
The limitation of this in silico research was that the data generated are predictive of the properties of the drug substance against the test receptor. Further in vivo and in vitro studies by animal study and cell cultures are warranted to validate the in silico data obtained in this study [69–72]. In this way, the completeness of the data for the development of antimalarial candidates can be achieved.
Conclusion
G. atroviridis has antimalarial potential based on a molecular docking experiment of phytochemical compounds against several Pf3D7 proteins (apPOL, ERS, PMX, and cyt c2 DSD). Quercetin had the strongest binding affinity for apPOL, ERS, and PMX. Kaempferol was the most effective inhibitor of cyt c2 DSD. Binding affinity indicates the ability of the drug to bind to the receptor. The smaller the binding affinity the higher the affinity of the complex. Further in vivo and in vitro analyses are required to demonstrate the efficacy of G. atroviridis as an antimalarial agent.
Acknowledgments
We thank Jalan Tengah, Indonesia (www.jalantengah.site) for editing the manuscript.
Conflict of interest
The authors declare that there are no conflicts of interest.
Authors’ contribution
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved the final manuscript for publication.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics statement
This study involved the use of secondary data retrieved from open databases, so ethical approval was not required.
Funding
This study was funded by the Virtual Research Center for Bioinformatics and Biotechnology, Indonesia.
Disclosure
Underlying data are available upon requested from the corresponding author. All authors listed stated that there were no conflicts of interest within the publication or funding.
References
- Melamane P, Shetty S, Gulati D. A study of drug resistance in malaria. J Indian Acad Clin Med 2014;15(1):9-12.
- Tasman MZ, Arwati H, Hasanatuldhiyyah N, Wardhani P. The differences of parasitemia in Plasmodium berghei infected mice treated with extract of mango parasite leaves with artemisinin combination. Qanun Medika 2023;7(1):53-60. [DOI: 10.30651/jqm.v7i1.13882]
- World Health Organization. “WHO Malaria Report”. World malaria report 2022. New York City: World Health Organization; 2022. pp. 7-30.
- Rich S, Leendertz FH, Xu G, LeBreton M, Djoko CF, et al. The origin of malignant malaria. Proc Natl Acad Sci USA 2009;106(35):14902-7. [PMID: 19666593 DOI: 10.1073/pnas.0907740106]
- Bakhraibah AO. Malaria in modern day: a review article. Austr J Basic Appl Sci 2018;12(11):73-5. [DOI: 10.22587/ajbas.2018.12.11.15]
- Baingana FK, Bos ER. Changing patterns of disease and mortality in Sub-Saharan Africa: an overview. In: Jamison DT, Feachem X, Makgoba MW, Bos ER, Baigana FK, Hofman KJ, et al. editors. Diseases and mortality in sub-Saharan Africa. 2nd ed. Washington: International Bank for Reconstruction and Development, The World Bank; 2006.
- Bhatia R, Rastogi RM, Ortega L. Malaria successes and challenges in Asia. J Vector Borne Dis 2013;50(4):239-47. [PMID: 24499845]
- Ali F, Wali H, Jan S, Zia A, Aslam M, et al. Analysing the essential proteins set of Plasmodium falciparum PF3D7 for novel drug targets identification against malaria. Malar J 2021;20:335. [PMID: 34344361: DOI: 10.1186/s12936-021-03865-1]
- Bonilla JA, Bonilla TD, Yowell CA, Fujioka H, Dame JB. Critical roles for the digestive vacuole plasmepsins of Plasmodium falciparum in vacuolar function. Mol Microbiol 2007;65(1):64-75. [PMID: 17581121 DOI: 10.1111/j.1365-2958.2007.05768.x]
- Oladele TO, Bewaji CO, Sadiku JS. Drug target selection for malaria: molecular basis for the drug discovery process. Centrepoint J (Sci Ed) 2012;18(2):111-24.
- Dong Y, Wan J, Sun A, Deng Y, Chen M, et al. Genetic association between the Pfk13 gene mutation and artemisinin resistance phenotype in Plasmodium falciparum isolates from Yunnan Province, China. Malar J 2018;17(1):478. [PMID: 30563521 DOI: 10.1186/s12936-018-2619-4]
- Yuan H, Ma Q, Ye L, Piao G. The traditional medicine and modern medicine from natural products. Molecules 2016;21(5):559. [PMID: 27136524 DOI: 10.3390/molecules21050559]
- Muschietti L, Vila R, Filho VC, Setzer W. Tropical protozoan diseases: natural product drug discovery and development. Evid Based Complement Alternat Med 2013;2013:404250. [PMID: 24348698 DOI: 10.1155/2013/404250]
- Hamidon H, Susanti D, Taher M, Zakaria ZA. Garcinia atroviridis — a review on phytochemicals and pharmacological properties. Marmara Pharm J 2018;21(24530):38-47. [DOI: 10.12991/marupj.259879]
- Shahid M, Law D, Azfaralariff A, Mackeen MM, Chong TF, et al. Phytochemicals and biological activities of Garcinia atroviridis: a critical review. Toxics 2022;10:656. [PMID: 36355947 DOI: 10.3390/toxics10110656]
- Al-Mansoub MA, Asmawi MZ, Murugaiyah V. Effect of extraction solvents and plant parts used on the antihyperlipidemic and antioxidant effects of Garcinia atroviridis: a comparative study. J Sci Food Agri 2014;94(8):1552-58. [PMID: 24166055 DOI: 10.1002/jsfa.6456]
- Suwanmannee S, Kitisin T, Luplertlop N. In vitro screening of 10 edible Thai plants for potential antifungal properties. Evid Based Complement Alternat Med 2014;2014:138587. [PMID: 24516502 DOI: 10.1155/2014/138587]
- Mackeen MM, Mooi LY, Amran M, Mat N, Lajis NH, et al. Noncytotoxic and antitumour-promoting activities of garcinia acid esters from Garcinia atroviridis Griff. ex T. Anders (Guttiferae). Evid Based Complement Alternat Med 2012;2012:829814. [PMID: 22685487 DOI: 10.1155/2012/829814]
- Syamsudin , Dewi RM, Hernita S. Efek ekstrak daun asam gelugur (Garcinia atroviridis Griff TAnders) terhadap Plasmodium berghei pada mencit. Airlangga J Pharm. 2004;4(3):101-4.
- Kulsum K, Syahrul S, Hasbalah K, Balqis U. Phytocompounds of Nigella sativa seeds extract and their neuroprotective potential via EGR1 receptor inhibition: a molecular docking study. Narra J 2023;3(2):e173. [PMID: 38454971 DOI: 10.52225/narra.v3i2.173]
- Meng X-Y, Zhang H-X, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 2012;7(2):146-57. [PMID: 21534921 DOI: 10.2174/157340911795677602]
- Nugraha RYB, Faratisha IFD, Mardhiyyah K, Ariel DG, Putri FF, et al. Antimalarial properties of isoquinoline derivative from Streptomyces hygroscopicus subsp. Hygroscopicus: an in silico approach. BioMed Res Int 2020;2020:6135696. [PMID: 31993450 DOI: 10.1155/2020/6135696]
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol 2015;1263:243-50. [PMID: 25618350 DOI: 10.1007/978-1-4939-2269-7_19]
- Listyani P, Kharisma VD, Ansori ANM, Widyananda MH, Probojati RT, et al. In silico phytochemical compounds screening of Allium sativum targeting the Mpro of SARS-CoV-2. Pharmacogn J 2022;14(3):604-9. [DOI: 10.5530/pj.2022.14.78]
- Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today: Technol 2004;1(4):337-41. [DOI: 10.1016/j.ddtec.2004.11.007]
- Umar AH, Ratnadewi D, Rafi M, Sulistyaningsih YC, Hamim H, et al. Drug candidates and potential targets of Curculigo spp. compounds for treating diabetes mellitus based on network pharmacology, molecular docking and molecular dynamics simulation. J Biomol Struct Dyn 2023;41(17):8544-60. [PMID: 36300505 DOI: 10.1080/07391102.2022.2135597]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, et al. NCBI BLAST: a better web interface. Nucleic Acids Res 2008;36(WS):W5-W9. [PMID: 18440982 DOI: 10.1093/nar/gkn201]
- Hasan MA, Khan MA, Sharmin T, Mazumder MHH, Chowdhury AS. Identification of putative drug targets in vancomycin-resistant Staphylococcus aureus (VRSA) using computer aided protein data analysis. Gene 2016;575(1):132-43. [PMID: 26319513 DOI: 10.1016/j.gene.2015.08.044]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074-D1082. [PMID: 29126136 DOI: 10.1093/nar/gkx1037]
- Zainul R, Kharisma VD, Ciuputri P, Ansori ANM, Herdiansyah MA, et al. Antiretroviral activity from elderberry (Sambucus nigra L.) flowers againts HIV-2 infection via reverse transcriptase inhibition: a viroinformatics study. Healthc Low Resour Sett 2024;1(2024):1-12. [DOI: 10.4081/hls.2024.12047]
- Herdiansyah MA, Ansori ANM, Kharisma VD, Alifiansyah MRT, Anggraini D, et al. In silico study of cladosporol and its acyl derivatives as anti-breast cancer against alpha-estrogen receptor. Biosaintifika 2024;16(1):142-54. [DOI: 10.15294/biosaintifika.v15i1.949]
- Shivanika C, Kumar D, Ragunathan V, Tiwari P, A S, P BD. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J Biomol Struct Dyn 2020;40(2):585-611. [PMID: 32897178 DOI: 10.1080/07391102.2020.1815584]
- Nandana PI, Rasyid H, Prihantono P, Yustitia I, Hakim L. Molecular docking studies of Brucein D as a potential inhibitor of the Bcl-2 anti-apoptotic protein. Bali Med J 2023;12(2):2148-52. [DOI: 10.15562/bmj.v12i2.4414]
- Krihariyani D, Haryanto E, Sasongkowati R. In silico analysis of antiviral activity and pharmacokinetic prediction of brazilein sappan wood (Caesalpinia sappan L.) against SARS-CoV-2 spike glycoproteins. Indonesian J Med Lab Sci Technol 2021;3(1):26-37. [DOI: 10.33086/ijmlst.v3i1.1854]
- Amrulloh LSWF, Harmastuti N, Prasetiyo A, Herowati R. Analysis of molecular docking and dynamics simulation of mahogany (Swietenia macrophylla King) compounds against the PLpro enzyme SARS-COV-2. Pharm Pharmaceut Sci J 2023;10(3):347-59. [DOI: 10.20473/jfiki.v10i32023.347-359]
- Chheda PR, Nieto N, Kaur S, Beck JM, Beck JR, et al. Promising antimalarials targeting apicoplast DNA polymerase from Plasmodium falciparum. Eur J Med Chem 2022;243:114751. [PMID: 36191407 DOI: 10.1016/j.ejmech.2022.114751]
- Sharma VK, Chhiber-Goel J, Yogavel M, Sharma A. Structural characterization of glutamyl-tRNA synthetase (GluRS) from Plasmodium falciparum. Mol Biochem Parasitol 2022;253:111530. [PMID: 36370911 DOI: 10.1016/j.molbiopara.2022.111530]
- Espino-Sanchez TJ, Wienkers H, Marvin RG, Nalder S-A, García-Guerrero AE, et al. Direct tests of cytochrome c and c1 functions in the electron transport chain of malaria parasites. Proc Natl Acad Sci USA 2023;120(19):e2301047120. [PMID: 37126705 DOI: 10.1073/pnas.2301047120]
- Kesari P, Deshmukh A, Pahelkar N, Suryawanshi AB, Rathore I, et al. Structures of plasmepsin X from Plasmodium falciparum reveal a novel inactivation mechanism of the zymogen and molecular basis for binding of inhibitors in mature enzyme. Pro Sci 2022;31:882-99. [PMID: 35048450 DOI: 10.1002/pro.4279]
- Thirumal Kumar T, Lavanya P, George Priya Doss C, Tayubi IA, Naveen Kumar DR, et al. A molecular docking and dynamics approach to screen potent inhibitors against fosfomycin resistant enzyme in clinical Klebsiella pneumoniae. J Cell Biochem 2017;118(11):4088-94. [PMID: 28409871 DOI: 10.1002/jcb.26064]
- Sneha P, Thirumal Kumar D, Tanwar H, Siva R, George Priya Doss C, Hatem Z. Structural analysis of G1691S variant in the human Filamin B gene responsible for Larsen syndrome: a comparative computational approach. J Cell Biochem 2017;118(7):1900-10. [PMID: 28145583 DOI: 10.1002/jcb.25920]
- Seeber F, Soldati-Favrez D. Metabolic pathways in the apicoplast of Apicomplexa. Int Rev Cell Mol Biol 2010;281:161-228. [PMID: 20460186 DOI: 10.1016/S1937-6448(10)81005-6]
- Yeh E, DeRisi JL. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol 2011;9(8):e1001138. [PMID: 21912516 DOI: 10.1371/journal.pbio.1001138]
- Padmi H, Kharisma VD, Ansori ANM, Sibero MT, Widyananda MH, et al. Macroalgae bioactive compounds for the potential antiviral of SARS-CoV-2: an in silico study. J Pure Appl Microbiol 2022;16(2):1018-27. [DOI: 10.22207/JPAM.16.2.26]
- Mailu BM, Ramasamay G, Mudeppa DG, Li L, Lindner SE, et al. A nondiscriminating glutamyl-tRNA synthetase in the Plasmodium apicoplast: the first enzyme in an indirect aminoacylation pathway. J Biol Chem 2013;288(45):32539-52. [PMID: 24072705 DOI: 10.1074/jbc.M113.507467]
- Manickam Y, Chaturvedi R, Babbar P, Malhotra N, Jain V, et al. Drug targeting of one or more aminoacyl-tRNA synthetase in the malaria parasite Plasmodium falciparum. Drug Discov Today 2018;23(6):1233-40. [PMID: 29408369 DOI: 10.1016/j.drudis.2018.01.050]
- Nachiappan M, Jain V, Sharma A, Yogavel M, Jeyakantham J. Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes. Int J Biol Macromol 2018;120(Pt B):1379-86. [PMID: 30248426 DOI: 10.1016/j.ijbiomac.2018.09.115]
- Pham JS, Dawson KL, Jackson KE, Lim EE, Pasaje CFA, et al. Aminoacyl-tRNA synthetases as drug targets in eukaryotic parasites. Int J Parasitol Drugs Drug Resist 2014;4(1):1-13. [PMID: 24596663 DOI: 10.1016/j.ijpddr.2013.10.001]
- Fang P, Han H, Wang J, Chen K, Chen X, et al. Structural basis for specific inhibition of tRNA synthetase by an ATP competitive inhibitor. Chem Biol 2015;22(6):734-44. [PMID: 26074468 DOI: 10.1016/j.chembiol.2015.05.007]
- Gora A, Brezovsky J, Damborsky J. Gates of enzymes. Chem Rev 2013;113(8):5871-923. [PMID: 23617803 DOI: 10.1021/cr300384w]
- Sharma VK, Gupta S, Chhiber-Goel J, Yogavel M, Sharma A. A single amino acid substitution alters activity and specificity in Plasmodium falciparum aspartyl & asparaginyl-tRNA synthetase. Mol Biochem Parasitol 2022;250:111488. [PMID: 35644266 DOI: 10.1016/j.molbiopara.2022.111488]
- Aini NS, Kharisma VD, Widyananda MH, Murtadlo AAA, Probojati RT, et al. Bioactive compounds from purslane (Portulaca oleracea L.) and star Anise (Illicium verum Hook) as SARS-CoV-2 antiviral agent via dual inhibitor mechanism: in silico approach. Pharmacogn J 2022;14(4):352-7. [DOI: 10.5530/pj.2022.14.106]
- Blasco B, Leroy D, Fidock DA. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med 2017;23(8):917-28. [PMID: 28777791 DOI: 10.1038/nm.4381]
- MacRae JI, Dixon MW, Dearnley MK, Chua HH, Chambers JM, et al. Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC Biol 2013;11:67. [PMID: 23763941 DOI: 10.1186/1741-7007-11-67]
- Pace CN, Fu H, Fryar KL, Landua J, Trevino SR, et al. Contribution of hydrophobic interactions to protein stability. J Mol Biol 2011;408(3):514-528. [PMID: 21377472 DOI: 10.1016/j.jmb.2011.02.053]
- Nasamu AS, Glushakova S, Russo I, Vaupel B, Oksman A, et al. Plasmepsins IX and X are essential and druggable mediators of malaria parasite egress and invasion. Science 2017;358(6362):518-22. [PMID: 29074774 DOI: 10.1126/science.aan1478]
- Pino P, Caldelari R, Mukherjee B, Vahokoski J, Klages N, et al. A multistage antimalarial targets the plasmepsins IX and X essential for invasion and egress. Science 2017;358(6362):522-8. [PMID: 29074775 DOI: 10.1126/science.aaf8675]
- Favuzza P, de Lera Ruiz M, Thompson JK, Triglia T, Ngo A, et al. Dual plasmepsin-targeting antimalarial agents disrupt multiple stages of the malaria parasite life cycle. Cell Host Microbe 2020;27(4):642-58. [PMID: 32109369 DOI: 10.1016/j.chom.2020.02.005]
- Sung S-S. Peptide folding driven by Van der Waals interactions. Pro Sci 2015;24:1383-8. [PMID: 26013298 DOI: 10.1002/pro.2710]
- Dahl EL, Rosenthal PJ. Multiple antibiotics exert delayed effects against the Plasmodium falciparum apicoplast. Antimicrob Agents Chemother 2007;51(10):3485-90. [PMID: 17698630 DOI: 10.1128/AAC.00527-07]
- Pradel G, Schlitzer M. Antibiotics in malaria therapy and their effect on the parasite apicoplast. Curr Mol Med 2010;10(3):335-49. [PMID: 20331433 DOI: 10.2174/156652410791065273]
- Antony HA, Parija SC. Antimalarial drug resistance: an overview. Trop Parasitol 2016;6(1):30-41. [PMID: 26998432 DOI: 10.4103/2229-5070.175081]
- Lim P, Chy S, Ariey F, Incardona S, Chim P, et al. Pfcrt polymorphism and chloroquine resistance in Plasmodium falciparum strains isolated in Cambodia. Antimicrob Agents Chemother 2003;47(1):87-94. [PMID: 12499174 DOI: 10.1128/AAC.47.1.87-94.2003]
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 2009;361(5):455-67. [PMID: 19641202 DOI: 10.1056/NEJMoa0808859]
- Anand David AV, Arulmoli R, Parasuraman S. Overviews of biological importance of quercetin: a bioactive flavonoid. Pharmacogn Rev 2016;10(20):84-9. [PMID: 28082789 DOI: 10.4103/0973-7847.194044]
- Yang D, Wang T, Long M, Li P. Quercetin: Its main pharmacological activity and potential application in clinical medicine. Oxid Med Cell Longev 2020;2020:8825387. [PMID: 33488935 DOI: 10.1155/2020/8825387]
- Periferakis A, Periferakis K, Badarau IA, Petran EM, Popa DC, et al. Kaempferol: antimicrobial properties, sources, clinical, and traditional applications. Int J Mol Sci 2022;23(23):15054. [PMID: 36499380 DOI: 10.3390/ijms232315054]
- Alam W, Khan H, Shah MA, Cauli O, Saso L. Kaempferol as a dietary anti-inflammatory agent: current therapeutic standing. Molecules 2020;25(18):4073. [PMID: 32906577 DOI: 10.3390/molecules25184073]
- Sugiharto S, Darmanto W, Wahyuningsih SPA, Hayati A, Martha Y, et al. Antioxidant activities of curcumin to MDA blood serum concentration and lead levels in liver of mice. Malaysian J Sci 2018;38(3):21-9. [DOI: 10.22452/mjs.sp2019no3.3]
- Sugiharto S, Winarni D, Islamatasya U, Bhakti IN, Nisa N, et al. Gynura procumbens adventitious root extract altered expression of antioxidant genes and exert hepatoprotective effects against cadmium-induced oxidative stress in mice. HAYATI J Biosci 2022;29(4):479-86. [DOI: 10.4308/hjb.29.4.479-486]
- Zubaidah U, Sugiharto S, Siregar MIP, Islamatasya U, Nisa N, et al. Gynura procumbens adventitious root ameliorates oxidative stress and has cytotoxic activity against cancer. BIOI 2024;5(1):1-9. [DOI: 10.15212/bioi-2024-0020]
- Kodariah L, Pakpahan SE, Aditya N, Nurzal ZR. Histopathological of Mice (Mus musculus) Liver Induced by Lead (Pb) Orally. Indones J Med Lab Sci Technol 2023;5(2):172-8. [DOI: 10.33086/ijmlst.v5i2.4295]
- Dweck AD. A Review of Asam Gelugor (Garcinia atroviridis) Griff ex T Anders. 1999. Available from: http://www.dweckdata.com/research_files/garcinia_atroviridis.pdf (accessed on 15 May 2024).
- Behera BC. Citric acid from Aspergillus niger: a comprehensive overview. Crit Rev Microbiol 2020;46(6):727-49. [PMID: 33044884 DOI: 10.1080/1040841X.2020.1828815]
- Kövilein A, Kubisch C, Cai L, Ochsenreither K. Malic acid production from renewables: a review. J Chem Technol Biotechnol 2020; 95(3):513-26. [DOI: 10.1002/jctb.6269]
- Kumar R, Basak B, Jeon B-H. Sustainable production and purification of succinic acid: a review of membrane-integrated green approach. J Clean Prod 2020;277:123954. [DOI: 10.1016/j.jclepro.2020.123954]
- Ismail A, Doghish AS, Elsadek BE, Salama SA, Mariee AD. Hydroxycitric acid potentiates the cytotoxic effect of tamoxifen in MCF-7 breast cancer cells through inhibition of ATP citrate lyase. Steroids 2020;160:108656. [PMID: 32439410 DOI: 10.1016/j.steroids.2020.108656]
- Li L, Chu X, Yao Y, Cao J, Li Q, et al. (-)-Hydroxycitric acid alleviates oleic acid-induced steatosis, oxidative stress, and inflammation in primary chicken hepatocytes by regulating amp-activated protein kinase-mediated reactive oxygen species levels. J Agric Food Chem 2020;68:11229-41. [DOI: 10.1021/acs.jafc.0c04648]
- Tomar M, Rao RP, Dorairaj P, Koshta A, Suresh S, et al. A clinical and computational study on anti-obesity effects of hydroxycitric acid. RSC Adv 2019;9(32):18578-88. [DOI: 10.1039/C9RA01345H]
- Galdiero E, Ricciardelli A, D’Angelo C, de Alteriis E, Maione A, et al. Pentadecanoic acid against Candida albicans–Klebsiella pneumoniae biofilm: towards the development of an anti-biofilm coating to prevent polymicrobial infections. Res Microbiol 2021;172(7-8):103880. [PMID: 34563667 DOI: 10.1016/j.resmic.2021.103880]
- To NB, Nguyen YTK, Moon JY, Ediriweera MK, Cho SK. Pentadecanoic acid, an odd-chain fatty acid, suppresses the stemness of MCF-7/SC human breast cancer stem-like cells through JAK2/STAT3 signaling. Nutrients 2020;12(6):1663. [PMID: 32503225 DOI: 10.3390/nu12061663]
- Fu W-C, Li H-Y, Li T-T, Yang K, Chen J-X, et al. Pentadecanoic acid promotes basal and insulin-stimulated glucose uptake in C2C12 myotubes. Food Nutr Res 2021;65:4527. [PMID: 33613155 DOI: 10.29219/fnr.v65.4527]
- Venn-Watson S, Lumpkin R, Dennis EA. Efficacy of dietary odd-chain saturated fatty acid pentadecanoic acid parallels broad associated health benefits in humans: Could it be essential? Sci. Rep 2020;10(1):8161. [PMID: 32424181 DOI: 10.1038/s41598-020-64960-y]
- Permana D, Lajis NH, Shaari K, Ali AM, Mackeen MM, et al. A new prenylated hydroquinone from the roots of Garcinia atroviridis Griff ex T. Anders (Guttiferae). Z Nat B 2003;58(4):332-5. [DOI: 10.1515/znb-2003-0414]
- Mackeen MM, Ali AM, Lajis NH, Kawazu K, Kikuzaki H, et al. Antifungal garcinia acid esters from the fruits of Garcinia atroviridis. Z Naturforschung C J Biosci 2002;57(3-4):291-5. [PMID: 12064729 DOI: 10.1515/znc-2002-3-416]
- Kosin J, Ruangrungsi N, Ito C, Furukawa H. A xanthone from Garcinia atroviridis. Phytochem 1998;47(6):1167-8. [DOI: 10.1016/S0031-9422(98)80095-2]
- Syahida A, Israf DA, Permana D, Lajis NH, Khozirah S, et al. Atrovirinone inhibits pro-inflammatory mediator release from murine macrophages and human whole blood. Immunol Cell Biol 2006;84(3):250-8. [PMID: 16509831 DOI: 10.1111/j.1440-1711.2006.01426.x]
- Permana D, Lajis NH, Mackeen MM, Ali AM, Aimi N, et al. Isolation and bioactivities of constitutents of the roots of Garcinia atroviridis. J Nat Prod 2001;64(7):976-9. [PMID: 11473441 DOI: 10.1021/np000563o]
- Tan W-N, Khairuddean M, Wong K-C, Khaw K-Y, Vikneswaran M. New cholinesterase inhibitors from Garcinia atroviridis. Fitoterapia 2014;97:261-7. [PMID: 24924287 DOI: 10.1016/j.fitote.2014.06.003]
- Permanaa D, Abas F, Maulidiani F, Shaari K, Stanslas J, et al. Atrovirisidone B, a new prenylated depsidone with cytotoxic property from the roots of Garcinia atroviridis. Z Naturforsch C J Biosci 2005;60(7-8):523-6. [PMID: 16163823 DOI: 10.1515/znc-2005-7-802]
- Tan W-N, Khairuddean M, Wong K-C, Tong W-Y, Ibrahim D. Antioxidant compounds from the stem bark of Garcinia atroviridis. J Asian Nat Prod Res 2016;18(8):804-11. [PMID: 26999039 DOI: 10.1080/10286020.2016.1160071]
- Tan W-N, Wong K-C, Khairuddean M, Eldeen IM, Asmawi MZ, Sulaiman B. Volatile constituents of the fruit of Garcinia atroviridis and their antibacterial and anti-inflammatory activities. Flavour Fragr J 2013;28(1):2-9. [DOI: 10.1002/ffj.3118]