Glycan-RNA: a new class of non-coding RNA
1Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
2Medical Research Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China
3Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria 3800, Australia
4Key Laboratory of Molecular Biology for Infectious Diseases, Ministry of Education, Institute for Viral Hepatitis, and Department of Infectious Diseases, the Second Affiliated Hospital, Chongqing Medical University, Chongqing, China
*Correspondence to: Yong Liao, E-mail: yongliao@cqmu.edu.cn; Phei Er Saw, E-mail: caipeie@mail.sysu.edu.cn
Received: October 15 2021; Revised: November 23 2021; Accepted: May 22 2022; Published Online: August 9 2022
Cite this paper:
Xiuling Li, Tiing Jen Loh, Jia Jia Lim, Phei Er Saw and Yong Liao. Glycan-RNA: a new class of non-coding RNA. BIO Integration 2022; 3(3): 124–131.
DOI: 10.15212/bioi-2021-0032. Available at: https://bio-integration.org/
Download citation
© 2022 The Authors. This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See https://bio-integration.org/copyright-and-permissions/
This commentary is based on a recent publication on the novel finding of a new class of non-coding RNA called glycan-RNA by Flynn et al.
Scientists have identified a new class of RNA functionalized with carbohydrates, which is present on cell surfaces. They have named this novel RNA glycoRNA according to its structure [1]. To better understand the characteristics of glycoRNA, we first describe basic information on RNA and glycosylation; we then discuss related topics on the identification and development of glycoRNAs.
RNAs
Ribonucleic acid (RNA), a type of nucleic acid, forms complex compounds with high molecular weight and takes deoxyribonucleic acid (DNA) as a template to produce protein while replacing DNA as a scaffold of genetic codes in some viruses, although RNA is limited to four bases [2, 3]. RNA functions primarily in cellular protein translation and synthesis, serving as a messenger carrying genetic information between DNA and ribosomes, helping ribosomes correctly assemble proteins, acting as biological catalysts, and participating in transcriptional and post-transcriptional genetic regulation [4, 5].
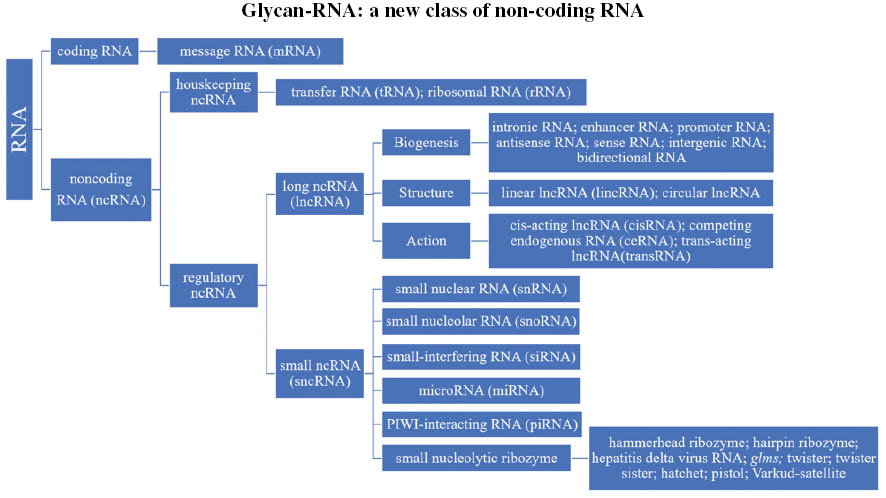
Of the many types of RNA, messenger RNA (mRNA), transfer RNA (tRNA) and ribosomal RNA (rRNA) are the three best known and most commonly studied. RNAs can be broadly classified into coding RNA and noncoding RNA (ncRNA). Two classes of ncRNAs exist—housekeeping ncRNAs (tRNA and rRNA) and regulatory ncRNAs—which are further divided according to size: those longer than 200 nucleotides are denoted long ncRNAs (lncRNA), whereas those shorter than 200 nucleotides are denoted small ncRNAs [6]. According to their biogenesis, structure and action, lncRNAs can be divided into many different classes (Figure 1) [7]. Small ncRNAs are subclassified into small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), small-interfering RNA (siRNA), microRNA (miRNA), PIWI-interacting RNA (piRNA) [8–10] and small nucleolytic ribozymes. These ribozymes are functional RNAs with enzymatic catalytic activity, which contain a loop or hairpin or hammerhead structure [11]. A large family of circular RNAs with self-cleaving functionality [12, 13] includes hammerhead ribozymes, hairpins, hepatitis delta virus RNA, glms, twister, twister sister, hatchet, pistol and Varkud-satellite [14, 15].
Figure 1 Tree of RNA systematics.
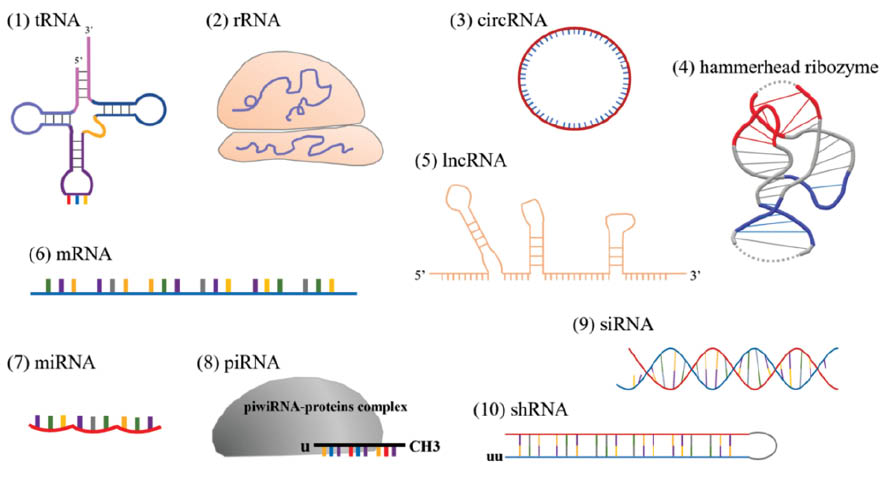
RNA-associated interactions broadly include RNA-RNA interactions, RNA-DNA interactions and RNA-protein interactions, which participate in a variety of cell pathological and physiological activities, such as cell growth, proliferation, differentiation and death [16]. RNA-RNA interactions include those between miRNA and mRNA, which result in mRNA degradation [17, 18]; those between tRNA and mRNA, which result in translation of genetic information [19]; and those involving RNA splicing, maturation and decay. Ribosomes are composed of rRNA and protein, and they interact with DNA in the transcription of the genetic code. RNA-binding proteins further participate in the regulation of gene expression and cellular functions [20]. Different types of RNA have unique structures that endow them with functionality (Figure 2).
Figure 2 Functional RNAs structures. (1) tRNA; (2) rRNA; (3) circRNA, circular RNA; (4) hammerhead ribozyme; (5) lncRNA; (6) mRNA; (7) miRNA; (8) piRNA; (9) siRNA; (10) shRNA, small hairpin ribozyme RNA.
In the field of transcriptomics, more than 140 types of RNA modification have been identified in four nucleobases [21]. In addition to the 5´ cap and 3´ poly(A) tail, which have been extensively researched, five primary classes of RNA modification have been identified: adenosine methylation, cytosine modifications, uridine isomerization, ribose modification and cytidine acetylation [22–24]. Functional RNAs can use base alterations to regulate their structure and function, and their modification is required for stability and proper biogenesis [25–31]. Through splicing, translation and decay, RNA modification ultimately modulates protein production [21], in a process called post-transcriptional modification. Beyond the chemical modification of ribose and nucleosides, RNA glycosylation [1] is a newly identified class of RNA modification, thus expanding the domain of epigenetics.
Glycosylation and glycans
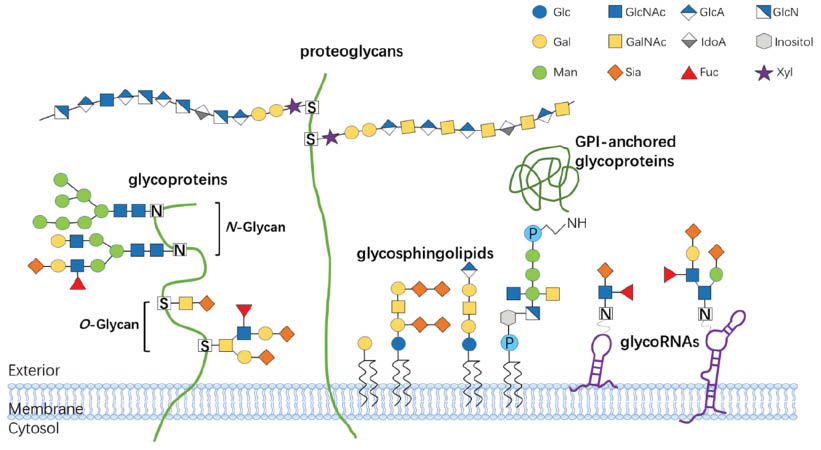
In glycosylation, a carbohydrate-like glycosyl donor is covalently attached to a hydroxyl or functional group of protein or lipid molecule, thereby forming a glycoconjugate through a series of enzymatic reactions in the Golgi apparatus and endoplasmic reticulum [32]. Glycosylation, the most complex post-translational modification of proteins and lipids, usually occurs at Ser/Thr residues (O-linked) or Asn-X-Ser/Thr consensus sequences (N-linked), thus forming a highly heterogeneous array of glycan structures on proteins or lipids [32, 33]. Glycoconjugates are formed from glycans and proteins or lipids though covalent bonding. According to their linkages with protein, lipid or glycan moieties, glycans are categorized into various of types. The major glycoconjugate types on the cell membrane are glycoproteins, proteoglycans, glycosphingolipids and glycosyl-phosphatidylinositol (GPI)-anchored glycoproteins [34, 35]. Among the N-linked glycans and O-linked glycans, protein glycosylation involves the addition of glycosaminoglycans (GAGs), phosphorylated glycans, C-mannosylated tryptophan residues and GPI anchors to peptide backbones [36]. Glycosphingolipids are formed through lipid glycosylation in the secretory pathway; they serve as components of plasma membranes and are functionally associated with the formation of lipid rafts [37] (Figure 3).
Figure 3 Major types of glycoconjugates on the cell membrane. Glycoproteins are a glycoconjugate class in which amino acid residues bind glycans and glycan chains via oxygen and nitrogen atoms of O-glycans and N-glycans. Proteoglycans are a class of glycoproteins characterized by long GAG chains with extended long disaccharide repeats (up to 50 disaccharide units) formed by GlcNAc or GalNAc linked to proteins. Glycosphingolipids consist of glycans and cellular membrane lipids. GPI-anchored glycoproteins are embedded in the outer plasma membrane and contain a glycan covalently attached to phosphatidylinositol. GlycoRNAs are a class of glycoconjugate in which glycans, such as Fuc or Sia, are conjugated to small conserved ncRNAs located at the cell surface. Geometric symbols depict types of sugars. Glc, glucose; GlcNAc, N-acetylglucosamine; GlcA, glucuronic acid; Gal, galactose; GalNAc, N-acetylgalactosamine; IdoA, iduronic acid; Man, mannose; Sia, sialic acid; Fuc, fucose; Xyl, xylose.
Glycans are branched molecules composed of monosaccharides assembled through chemical bonds. Glycans are synthesized from seven monosaccharides: fucose (Fuc), sialic acid (SA), glucose (Glc), galactose (Gal), mannose (Man), N-acetylglucosamine (GlcNAc) and N-acetyl-galactosamine (GalNAc). Fuc and SA are usually found at the termini of glycosylated glycan chains [38]. In fucosylation and sialylation, Fuc and SA, respectively, are attached to a functional group or the hydroxyl of a protein.
Glycosylation enhances protein stabilization destabilizing unfolded proteins, on the basis of thermodynamic analysis [39]. N-glycosylation helps protein fold into the correct three-dimensional form to enable biological functions, such as cell-cell interactions and cell signaling [40, 41]. N-linked glycosylation can prevent the deamidation of glycoproteins and glycopeptides—a common pathway of protein degradation [42]. Glycosylated proteins are more stable than non-glycosylated proteins, owing to longer half-lives after deamidation. The mechanism by which glycosylation arrest proteins deamidation is that large N-glycans sterically hinder attack of the amine groups of side chains by backbone nitrogen, thereby preventing formation of a hydrolyzable succinimide intermediate that result in hydrolysis [43, 44]. Nascent glycoproteins bind membrane-bound calnexin and soluble calreticulin, two endoplasmic reticulum chaperones, which retain correctly unfolded proteins in the endoplasmic reticulum until the terminal glucose residue of the sugar chain is eliminated by glucosidase, and glycoprotein assembly is correctly completed [45, 46]. Protein glycosylation influences protein folding, degradation and trafficking to the proper destinations, such as targeting of receptors to the cell surface [47].
Glycosylation can alter the interactions between ligands and receptors, thus regulating signal transduction [35]. Sialylation is a switch-off signal of integrin glycan receptor combining with endogenous lectin galectin-1 (Gal-1) ligand, which may enable tumor cells to develop Gal-1 dependent anoikis resistance [48]. Obesity-induced insulin resistance is associated with the activity of FcγRIIB, an endothelial immunoglobulin G (IgG) receptor, which can be invalidated by sialylated IgG [49]. Glycosylation-dependent cell adhesion molecules are members of the L-selectin ligand family, whose sialylation and fucosylation modifications stimulate lymphocyte homing through tethering and rolling adhesion, as well as the integrin-activation pathway [50]. Blood-group antigens, the terminal epitopes generated by sialic acid and fucose residues on the sugar chains, are glycans conjugated with proteins [51, 52], thus indicating that glycans can be antigenic.
As immune system effectors, glycol forms are key in the synthesis, stability, signal recognition, function regulation and protein-protein interaction of immune proteins [53, 54]. The unassembled heavy chains of major histocompatibility complex I (MHC I) interact with membrane bound calnexin, and Asn residues must be modified with sugar chains for initial assembly. The transmembrane glycoprotein tapasin acts as a scaffold integrating a complex of MHC I, calreticulin, ERp57 and TAP transporter, thus forming mature MHC I [55, 56]. The integration of antigenic glycopeptides on peptide moieties and glycans defines the specificity of T cells [57]. Through the classical MHC relative way, monosaccharide and disaccharide alterations of glycopeptides are recognized by CD8+ and CD4+ T cells [58, 59].
Glycosylation modifications of proteins on the cell surface have been found to be associated with multidrug resistance [60]. In acute myeloid leukemia (AML) mouse models, inflammatory mediators, such as TNF-α and G-CSF [61, 62] released from AML cells alter the glycosylation of cell-surface ligand sugar chains by coordinating the activities of sialyltransferase and fucosyltransferase. Consequently, AML cell binding to endothelial E-selectin receptors [63] may cause chemotherapy resistance [64] through altering the endothelial niche microenvironment and activating the AKT/NF-κB/mTOR pathway. The expression of relative glycosylases is associated with DNA methylation and histone modulation [65, 66]. CD63 is a protein that can recruit or link receptor tyrosine kinases to integrins and Src family kinases, thus resulting in the development of cancer malignancy [67] via the N-glycosylation modification. Glycosylated CD63 is transported to the cell surface, where it has functions in invasion and drug resistance of breast cancer cells, whereas non-glycosylated CD63 remains trapped in the endosomal system, thus resulting in tumor cell non-malignancy [68]. N-glycan is a major component of the epidermal growth factor receptor (EGFR), which can regulate its cell surface expression and cause a drug resistance phenotype to EGFR inhibition [69]. N-glycosylated P-glycoprotein is more stable than non-glycosylated P-glycoprotein, and consequently is anchored to the cell membrane and promotes multidrug resistance [70–72].
Aberrant glycosylation of proteins may lead to disintegration of the cell membrane. In Aspergillus, the abnormal synthesis and transport of GDP-mannose, a mannosylation precursor, modulates cell viability, cell antigen phenotypes and cell membrane integrity by altering the function of GDP-mannose transporters and related catalytic enzymes [73, 74].
Glycosylation often occurs on proteins, lipids and glycans [34, 35], whereas RNA is usually not considered a major target of glycosylation. However, a new study has recently found that mammalian cell types and animals use RNA as a third scaffold substrate for glycosylation (Figure 3).
GlycoRNA: glycans are directly linked to RNA
Research has indicated a new biological phenomenon in the RNA field in which conserved small ncRNAs have been found to bear N-glycans. The term glycoRNAs has been coined to describe those small highly sialylated and fucosylated ncRNAs at the cell surface [31]. GlycoRNAs are a new breakthrough in the fields of RNA and glycobiology. GlycoRNA assembly depends on the canonical N-glycan biosynthetic machinery, which catalyzes sialic acid and fucose enrichment. These RNAs are localized on cell surface, where they interact with anti-dsRNA antibodies and members of the sialic acid-binding immunoglobulin-type lectins (Siglec) receptor family. GlycoRNAs are a class of small ncRNAs with a common set of transcripts, such as Y RNA, snRNA, rRNA, snoRNA and tRNA. The Y5 RNA transcript is strongly enriched in the pool of candidate glycoRNAs.
Y5 RNA is a member of family Y ncRNAs in human genes with a highly conserved stem-loop structure [75, 76]; it is transcribed by RNA polymerase III [77], and it frequently binds Ro60 protein, La protein or their orthologs in a loop-dependent manner [78, 79]. The localization of Y5 RNA is mostly nuclear [80, 81], and its transport to the cytoplasm is triggered by Ro60-dependent nuclear export, a lack of La-binding to the 3´ end of Y RNA or its trimming [82, 83]. Other classes of glycoRNA transcripts, such as sn/snoRNAs, are localized to the cytoplasm and nucleus, respectively. Flynn et al. [31] have discovered that glycoRNAs are associated with the cellular membrane but not the soluble cytosol or nucleus, thus suggesting that interactions with glycan moieties on RNA transcripts might influence RNA localization and lead to yet-unknown functional changes.
Evidence indicates that glycoRNA glycans are structurally related to those found on proteins. Thus, study of the transferases participating in protein glycosylation may clarify the associations between RNA and glycoRNA-associated glycans. Glycan structures on glycoRNA have been defined as N-glycans, and only N-glycosylation enzymes can regulate the biosynthesis of glycoRNA. An apparent dose-dependent loss of glycoRNA label would occur after treatment with endoglycosidases, which are highly selective for N-glycans, and small molecule inhibitors of N-glycan trimming enzymes. Partial loss of label or even no effect has been observed after treatment with weakly selective N-glycan digesting enzymes or other enzymes, such as O-glycosidase and mucinase. Glycosylation-associated enzymes have been found to mediate glyco-complex biosynthesis in both the RNA and protein fields. Glycosyltransferase-encoding genes are epigenetically regulated by DNA methylation and histone acetylation, and produce specific type N-glycans in the proteome [84–86]. DNA methylation of genes also shapes the subclasses of IgG by modulating IgG glycan synthesis [87]. Because glycans have high homology, on the basis of proteomics and RNA nucleic omics, DNA methylation and histone acetylation can rationally be speculated to tune the biosynthesis of glycoRNA through mechanisms similar to those of glycoproteins.
The team used an azide-labeled precursor of sialic acid, peracetylated N-azidoacetylmannosamine (Ac4ManNAz), as an azidosugar to label living cells; they found that azide reactivity was focused on highly purified RNA preparations rather than other cell lysates [31]. The team had established methods based on metabolic labeling and biorthogonal chemistry to study protein-associated glycans several years prior [88–90]. The preliminary work was based on the hypothesis that labels with precursor sugars decorated with an azide group, called azidosugars, would be incorporated into cellular glycans and would undergo a biorthogonal reaction with biotin probes in cells or animals. Because biorthogonal reactions occur between azidosugars and cellular glycans, and because azide reactivity was concentrated on RNA preparations, the authors speculated that RNA might potentially interact with cellular glycans.
The authors confirmed that the RNA, but not other nucleotides, such as DNA, extracted from the labeled cells showed biotin reactivity, which was reversed by RNase treatment. GlycoRNAs with N-glycan decoration are highly fucosylated and sialylated. Because the study was limited to labeling of one glycan (sialic acid), RNAs modified with sialoglycans might constitute only one class of glycoRNA, whereas not all glycans, decorating with sialic acid and other glycol-forms, may be able to be conjugated to RNAs. How the RNA template conjugates to carbohydrate and why RNA can localize to the cell membrane are further questions to be explored.
Glycosylation-associated ncRNA vs. glycoRNA
Before identification of this new class of ncRNA, the relationship between RNA and glycans was known: ncRNA regulates glycosylation of proteins and the function of glycosyltransferase, which remodels glycans and influences cell activity. Protein glycosylation is a primary post-translational modification that substantially influences protein folding, localization, stability and activity. Ranging from simple monosaccharide modifications of nuclear transcription factors to highly complex branched polysaccharide modification of cell-surface receptors, glycosylation encompasses diverse sugar addition to proteins.
N-acetylgalactosamine transferase (GALNT) is an enzyme that initiates the cascade of mucin type O-linked glycosylation, whose presence at the cell surface can lead to metabolic disorders and cancers [91]. Li et al. [92] have found that the lncRNA SNHG7 acts as a competing endogenous RNA that sponges miR-34a, thus blocking binding of GALNT and miR-34a. Without the limitation of miR-34a, GALNT expression in cancer tissues is strengthened, thus leading to cancer proliferation, invasion and metastasis in the context of aberrant O-glycosylation [93].
Specific fucosyltransferases (FUTs) play major roles in malignant cancer processes by catalyzing aberrant fucosylation. Xu et al. [94] have confirmed that the exosome-derived lncRNA MALAT1 directly competes for miR-26a/26b binding sites and increases FUT4 expression and fucosylation levels, thus promoting metastasis of colorectal cancer. The lncRNA HOTAIR, associated with poor clinical prognosis, sponges miR-326 and consequently regulates FUT levels by modifying the fucosylation of the E-selectin ligand [95]. Some microRNAs can also regulate other members of the FUT family, thereby altering glycan production [96, 97].
Being the substrate of sialyltransferase ST6GAL1, sialylation of activated epidermal growth factor receptor could be mediated and regulated by the ZFAS1/miR-150/ST6GAL1 axis, thus conferring a multi-drug resistance phenotype via the activated PI3K/AKT pathway [98]. ST6GAL1 and the lncRNA HOTAIR are direct targets of miR-214, and HOTAIR regulates the expression of ST6GAL1 by sponging miR-214 [99]. ST6GAL1 leads to metabolic sialylation of c-Met though the JAK2/STAT3 pathway, thus promoting colorectal cancer malignancy. Sialyltransferase family members interact with miRNA by altering glycosylation patterns, thus affecting the progression of breast cancer [96].
The newly discovered glycoRNAs differ from glycosylation-associated ncRNAs. Glycosylation-associated ncRNAs regulate glycan-associated protein expression and alter glycosylation patterns, thus inducing disease occurrence. These sugars are not directly linked to ncRNA under the reaction between ncRNA and related proteins. GlycoRNAs are a new class of ncRNA in which RNA is directly linked with glycan; the glycoRNA carries genetic information and is present on the cell surface, thus distinguishing it from glycosylation-associated ncRNA.
In glycosylation, glycans decorate other biological polymers, thus allowing cells to construct extensive molecular forms from the same DNA blueprint. Scientists had long believed that only proteins and lipids can be linked to carbohydrates. However, new research from Flynn R.A. et al. has indicated that RNA can also be glycosylated, and the glycosylation-modified nucleic acids are located on cell surfaces. This report has provided the first observation of this feature in the RNA field.
GlycoRNAs are located on the outer cell membrane, where they bind Siglecs; therefore, glycoRNAs may have roles in immune signal transduction. Siglecs are an immune receptor family associated with various of diseases, such as systemic lupus erythematosus. Because of their ability to interact with anti-dsRNA antibodies and Siglec receptors, glycoRNAs may become a new serum marker of several diseases, thus enabling rapid clinical disease diagnosis. RNA modified with glycans may be sensitive to immunotherapy medicines [60] and consequently may serve as drug targets to increase drug response and drug enrichment in lesions. Immunotherapy, radiotherapy, chemotherapy and other methods may be used to alter the genetic information or glycan structure of glycoRNAs on the cell surface, thus therapeutically altering the lesion microenvironment. GlycoRNAs are expected to become a new type of immune response signal receiver, beyond protein receptors, in drug therapy.
Intriguingly, an emerging paradigm suggests that synthetic and clickable sugars that label glycoproteins and glycolipids can also modify small noncoding RNAs, thus making RNA the third scaffold for glycosylation. The first evidence indicating that highly sialylated and fucosylated glycoRNAs (RNA-glycan conjugates) are displayed on the cell surface, and can bind Siglec receptors and be decorated with N-glycans, has elevated studies of RNA biology, glycobiology and the glycome (particularly the sialome and fucosylome) to a new level of complexity. Although understanding and deciphering of the chemical structures and molecular functions of specific glycoRNAs on the cell surface remain limited, the discovery of glycoRNAs in mammalian cells may enable wider use and integration of glycoRNA data in traditional glycomics and omics workflows in cell biology, and enhance understanding of human diseases, thus enabling the design of novel glycan-based natural and glycoRNA mimetic therapeutics in the future.
References
- Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 2021;184(12):3109-24 e22. [PMID: 34004145 DOI: 10.1016/j.cell.2021.04.023]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157(1):77-94. [PMID: 24679528 DOI: 10.1016/j.cell.2014.03.008]
- Sharp PA. The centrality of RNA. Cell 2009;136(4):577-80. [PMID: 19239877 DOI: 10.1016/j.cell.2009.02.007]
- Saw PE, Song EW. siRNA therapeutics: a clinical reality. Sci China Life Sci 2020;63(4):485-500. [PMID: 31054052 DOI: 10.1007/s11427-018-9438-y]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43(6):904-14. [PMID: 24678707 DOI: 10.2174/1568026614666140329230311]
- Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol 2016;937:3-17. [PMID: 27573892 DOI: 10.1007/978-3-319-42059-2_1]
- Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, et al. Long non-coding RNA: classification, biogenesis and functions in blood cells. Mol Immunol 2019;112:82-92. [PMID: 31079005 DOI: 10.1016/j.molimm.2019.04.011]
- Crooke ST, Liang XH, Baker BF, Crooke RM. Antisense technology: a review. J Biol Chem. 2021;296:100416. [PMID: 30446728 DOI: 10.1016/j.jbc.2021.100416]
- Liu Y, Dou M, Song X, Dong Y, Liu S, et al. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer 2019;18(1):123. [DOI: 10.1186/s12943-019-1052-9]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 2019;20(2):89-108. [PMID: 30446728 DOI: 10.1038/s41576-018-0073-3]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, et al . Self-splicing RNA Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 1982;31(1):147-57. [PMID: 6297745 DOI: 10.1016/0092-8674(82)90414-7]
- de la Pena M, Garcia-Robles I, Cervera A. The Hammerhead Ribozyme: A Long History for a Short RNA. Molecules 2017;22(1):78. [PMID: 28054987 DOI: 10.3390/molecules22010078]
- Seith DD, Bingaman JL, Veenis AJ, Button AC, Bevilacqua PC. Elucidation of Catalytic Strategies of Small Nucleolytic Ribozymes From Comparative Analysis of Active Sites. ACS Catal 2018;8(1):314-27. [PMID: 32547833 DOI: 10.1021/acscatal.7b02976]
- Jimenez RM, Polanco JA, Luptak A. Chemistry and Biology of Self-Cleaving Ribozymes. Trends Biochem Sci 2015;40(11):648-61. [PMID: 26481500 DOI: 10.1016/j.tibs.2015.09.001]
- Ward WL, Plakos K, DeRose VJ. Nucleic acid catalysis: metals, nucleobases, and other cofactors. Chem Rev 2014;114(8):4318-42. [PMID: 24730975 DOI: 10.1021/cr400476k]
- Lin Y, Liu T, Cui T, Wang Z, Zhang Y, et al. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res 2020;48(D1):D189-D97. [PMID: 31906603 DOI: 10.1093/nar/gkz804]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [PMID: 15372042 DOI: 10.1038/nature02871]
- Bartel DP. MicroRNAs genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [PMID: 14744438 DOI: 10.1016/s0092-8674(04)00045-5]
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, et al. Structure of a ribonucleic acid. Science 1965;147:1462-5. [PMID: 14263761 DOI: 10.1126/science.147.3664.1462]
- Perez-Perri JI, Rogell B, Schwarzl T, Stein F, Zhou Y, et al. Discovery of RNA-binding proteins and characterization of their dynamic responses by enhanced RNA interactome capture. Nat Commun 2018;9(1):4408. [PMID: 30352994 DOI: 10.1038/s41467-018-06557-8]
- Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, et al. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res 2006;34(Database issue):D145-9. [PMID: 29106616 DOI: 10.1093/nar/gkx1030]
- Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell 2018;175(7):1872-86 e24. [PMID: 30449621 DOI: 10.1016/j.cell.2018.10.030]
- Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017;169(7):1187-200. [PMID: 28622506 DOI: 10.1016/j.cell.2017.05.045]
- Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods 2016;14(1):2331-200. [PMID: 28032622 DOI: 10.1038/nmeth.4110]
- Hoernes TP, Clementi N, Faserl K, Glasner H, Breuker K, et al. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res 2016;44(2):852-62. [PMID: 26578598 DOI: 10.1093/nar/gkv1182]
- Yildirim I, Kierzek E, Kierzek R, Schatz GC. Interplay of LNA and 2’-O-methyl RNA in the structure and thermodynamics of RNA hybrid systems: a molecular dynamics study using the revised AMBER force field and comparison with experimental results. J Phys Chem B 2014;118(49):14177-87. [PMID: 25268896 DOI: 10.1021/jp506703g]
- Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 2000;49(5):341-51. [PMID: 10902565 DOI: 10.1080/152165400410182]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 2012;40(11):5023-33. [PMID: 22344696 DOI: 10.1093/nar/gks144]
- Li X, Xiong X, Wang K, Wang L, Shu X, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol 2016;12(5):311-6. [PMID: 26863410 DOI: 10.1038/nchembio.2040]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016;530(7591):441-6. [PMID: 26863196 DOI: 10.1038/nature16998]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, et al. Erratum: Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015;527(7577):264. [PMID: 26416736 DOI: 10.1038/nature15717]
- Stanley P. Golgi glycosylation. Cold Spring Harb Perspect Biol 2011;3(4):a005199. [PMID: 21441588 DOI: 10.1101/cshperspect.a005199]
- Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, et al. N-Linked glycosylation in campylobacter jejuni and its functional transfer into E. coli. Science 2002;298(5599):1790-3. [PMID: 12459590 DOI: 10.1126/science.298.5599.1790]
- Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer 2005;5(7):526-42. [PMID: 16069816 DOI: 10.1038/nrc1649]
- Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol 2019;15(6):346-66. [PMID: 30858582 DOI: 10.1038/s41581-019-0129-4]
- Spiro RG. Protein glycosylation nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002;12(4):43-56. [PMID: 12042244 DOI: 10.1093/glycob/12.4.43r]
- Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 1999;9(8):747-55. [PMID: 10406840 DOI: 10.1093/glycob/9.8.747]
- Brooks SA. Appropriate Glycosylation Of Recombinant proteins for human use. Mol Biotechnol 2004;28:241-55. [PMID: 15542924 DOI: 10.1385/MB:28:3:241]
- Shental-Bechor D, Levy Y. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci USA 2008;105(24):8256-61. [PMID: 18550810 DOI: 10.1073/pnas.0801340105]
- Shental-Bechor D, Levy Y. Folding of glycoproteins: toward understanding the biophysics of the glycosylation code. Curr Opin Struct Biol 2009;19(5):524-33. [PMID: 19647993 DOI: 10.1016/j.sbi.2009.07.002]
- Varki A. Biological roles of oligosaccharides all of the theories are correct. Glycobiology 1993;2(3):97-130. [PMID: 8490246 DOI: 10.1093/glycob/3.2.97]
- Zhu HJ, Liu D, Tran VP, Wu Z, Jiang K, et al. N-Linked glycosylation prevents deamidation of glycopeptide and glycoprotein. ACS Chem Biol 2020;15(12):3197-205. [DOI: 10.1021/acschembio.0c00734]
- McFadden PN, Clarke S. Conversion of isoaspartyl peptides to normal peptides implications for the cellular repair of damaged proteins. Proc Natl Acad Sci USA 1987;84(9):2595-9. [PMID: 3472227 DOI: 10.1073/pnas.84.9.2595]
- Tyler-Cross R, Schirch V. Effects of amino acid sequence, buffers, and ionic strength on the rate and mechanism of deamidation of asparagine residues in small peptides. J Biol Chem 1991;266(33):22549-56. [PMID: 1939272]
- Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, et al . The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem 1999;274(6):3453-60. [PMID: 9920890 DOI: 10.1074/jbc.274.6.3453]
- Werner ED, Brodsky JL, McCracken AA. Proteasome-dependent endoplasmic reticulum-associated protein degradation An unconventional route to a familiar fate. Proc Natl Acad Sci USA 1996;93(24):13797-801. [PMID: 8943015 DOI: 10.1073/pnas.93.24.13797]
- Dove A. The bittersweet promise of glycobiology. Nature Biotechnology 2001;19:913-7. [PMID: 11581651 DOI: 10.1038/nbt1001-913]
- Sanchez-Ruderisch H, Detjen KM, Welzel M, Andre S, Fischer C, et al. Galectin-1 sensitizes carcinoma cells to anoikis via the fibronectin receptor alpha5beta1-integrin. Cell Death Differ 2011;18(5):806-16. [PMID: 21113146 DOI: 10.1038/cdd.2010.148]
- Tanigaki K, Sacharidou A, Peng J, Chambliss KL, Yuhanna IS, et al. Hyposialylated IgG activates endothelial IgG receptor FcgammaRIIB to promote obesity-induced insulin resistance. J Clin Invest 2018;128(1):309-22. [PMID: 29202472 DOI: 10.1172/JCI89333]
- Tangemann K, Bistrup A, Hemmerich S, Rosen SD. Sulfation of a high endothelial venule-expressed ligand for L-selectin. Effects on tethering and rolling of lymphocytes. J Exp Med 1999;190(7):935-41. [PMID: 10510083 DOI: 10.1084/jem.190.7.935]
- Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol 2008;1(3):183-97. [PMID: 19079178 DOI: 10.1038/mi.2008.5]
- Vitiazeva V, Kattla JJ, Flowers SA, Linden SK, Premaratne P, et al. The O-Linked Glycome and Blood Group Antigens ABO on Mucin-Type Glycoproteins in Mucinous and Serous Epithelial Ovarian Tumors. PLoS One 2015;10(6):e0130197. [PMID: 26075384 DOI: 10.1371/journal.pone.0130197]
- Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science 2001;291(5512):2370-6. [PMID: 11269318 DOI: 10.1126/science.291.5512.2370]
- Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem 2003;72:643-91. [PMID: 12676797 DOI: 10.1146/annurev.biochem.72.121801.161809]
- Leeuwen JEMV, Kearse KP. Deglucosylation of N-linked glycans is an important step in the dissociation of calreticulin–class I–TAP complexes. Proc Natl Acad Sci USA 1996;93(24):13997-4001. [PMID: 8943049 DOI: 10.1073/pnas.93.24.13997]
- Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity 1996;5(2):103-14. [PMID: 8769474 DOI: 10.1016/s1074-7613(00)80487-2]
- Glithero A, Tormo J, Haurum JS, Arsequell G, Valencia G, et al. Crystal structures of two H-2DbGlycopeptide complexes suggest a molecular basis for CTL cross-reactivity. Immunity 1999;10(1):63-74. [PMID: 10023771 DOI: 10.1016/s1074-7613(00)80007-2]
- Abdel-Motal UM, Berg L, Rosén A, Bengtsson M, Thorpe CJ, et al. Immunization with glycosylated Kb-binding peptides generates carbohydrate-specific, unrestricted cytotoxic T cells. Eur J Immunol 1996;26(3):544-51. [PMID: 8605919 DOI: 10.1002/eji.1830260307]
- Hudrisier D, Riond J, Mazarguil H, Oldstone MB, Gairin JE. Genetically encoded and post-translationally modified forms of a major histocompatibility complex class I-restricted antigen bearing a glycosylation motif are independently processed and co-presented to cytotoxic T lymphocytes. J Biol Chem 1999;274(51):36274-80. [PMID: 10593916 DOI: 10.1074/jbc.274.51.36274]
- Ma H, Zhou H, Song X, Shi S, Zhang J, et al . Modification of sialylation is associated with multidrug resistance in human acute myeloid leukemia. Oncogene 2015;34(6):726-40. [PMID: 24531716 DOI: 10.1038/onc.2014.7]
- Dagia NM, Gadhoum SZ, Knoblauch CA, Spencer JA, Zamiri P, et al. G-CSF induces E-selectin ligand expression on human myeloid cells. Nat Med 2006;12(10):1185-90. [PMID: 16980970 DOI: 10.1038/nm1470]
- Delmotte P, Degroote S, Lafitte JJ, Lamblin G, Perini JM, et al. Tumor necrosis factor alpha increases the expression of glycosyltransferases and sulfotransferases responsible for the biosynthesis of sialylated and/or sulfated Lewis x epitopes in the human bronchial mucosa. J Biol Chem 2002;277(1):424-31. [PMID: 11679593 DOI: 10.1074/jbc.M109958200]
- Mondal N, Buffone A Jr., Stolfa G, Antonopoulos A, Lau JT, et al. ST3Gal-4 is the primary sialyltransferase regulating the synthesis of E-, P-, and L-selectin ligands on human myeloid leukocytes. Blood 2015;125(4):687-96. [PMID: 25498912 DOI: 10.1182/blood-2014-07-588590]
- Barbier V, Erbani J, Fiveash C, Davies JM, Tay J, et al. Endothelial E-selectin inhibition improves acute myeloid leukaemia therapy by disrupting vascular niche-mediated chemoresistance. Nat Commun 2020;11(1):2042. [PMID: 32341362 DOI: 10.1038/s41467-020-15817-5]
- Pink M, Ratsch BA, Mardahl M, Durek P, Polansky JK, et al. Imprinting of skin/inflammation homing in CD4(+) T cells is controlled by DNA methylation within the fucosyltransferase 7 gene. J Immunol 2016;197(8):3406-14. [PMID: 27591321 DOI: 10.4049/jimmunol.1502434]
- Syrbe U, Jennrich S, Schottelius A, Richter A, Radbruch A, et al . Differential regulation of P-selectin ligand expression in naive versus memory CD4+ T cells: evidence for epigenetic regulation of involved glycosyltransferase genes. Blood 2004;104(10):3243-8. [PMID: 15297307 DOI: 10.1182/blood-2003-09-3047]
- Berditchevski F, Tolias KF, Wong K, Carpenter CL, Hemler ME. A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J Biol Chem 1997;272(5):2595-8. [PMID: 9006891 DOI: 10.1074/jbc.272.5.2595]
- Tominaga N, Hagiwara K, Kosaka N, Honma K, Nakagama H, et al. RPN2-mediated glycosylation of tetraspanin CD63 regulates breast cancer cell malignancy. Mol Cancer 2014;13(134):134. [PMID: 24884960 DOI: 10.1186/1476-4598-13-134]
- Contessa JN, Bhojani MS, Freeze HH, Ross BD, Rehemtulla A, et al . Molecular imaging of N-linked glycosylation suggests glycan biosynthesis is a novel target for cancer therapy. Clin Cancer Res 2010;16(12):3205-14. [PMID: 20413434 DOI: 10.1158/1078-0432.CCR-09-3331]
- Honma K, Iwao-Koizumi K, Takeshita F, Yamamoto Y, Yoshida T, et al. RPN2 gene confers docetaxel resistance in breast cancer. Nat Med 2008;14(9):939-48. [PMID: 18724378 DOI: 10.1038/nm.1858]
- Kramer R, Weber TK, Arceci R, Ramchurren N, Kastrinakis WV, et al. Inhibition of N-linked glycosylation of P-glycoprotein by tunicamycin results in a reduced multidrug resistance phenotype. Br J Cancer 1995;71(4):670-5. [PMID: 7710927 DOI: 10.1038/bjc.1995.133]
- Schinkel AH, Kemp S, Dollé M, Rudenko G, Wagenaar E. N-glycosylation and deletion mutants of the human MDR1 P-glycoprotein. J Biol Chem 1993;268(10):7474-81. [PMID: 8096511]
- Jiang H, Ouyang H, Zhou H, Jin C. GDP-mannose pyrophosphorylase is essential for cell wall integrity, morphogenesis and viability of Aspergillus fumigatus. Microbiology (Reading) 2008;154(Pt 9):2730-9. [PMID: 18757806 DOI: 10.1099/mic.0.2008/019240-0]
- Kadry AA, El-Ganiny AM, Mosbah RA, Kaminskyj SGW. Deletion of Aspergillus nidulans GDP-mannose transporters affects hyphal morphometry, cell wall architecture, spore surface character, cell adhesion, and biofilm formation. Med Mycol 2018;56(5):621-30. [DOI: 10.1093/mmy/myx082]
- Kohn M, Pazaitis N, Huttelmaier S. Why YRNAs? About versatile RNAs and their functions. Biomolecules 2013;3(1):143-56. [PMID: 24970161 DOI: 10.3390/biom3010143]
- Shi J, Zhou T, Chen Q. Exploring the expanding universe of small RNAs. Nat Cell Biol 2022;24(4):415-23. [PMID: 35414016 DOI: 10.1038/s41556-022-00880-5]
- Boire G, Craft J. Human Ro ribonucleoprotein particles: characterization of native structure and stable association with the La polypeptide. J Clin Invest 1990;85(4):1182-90. [PMID: 1690756 DOI: 10.1172/JCI114551]
- O’Brien CA, Harley JB. A subset of hY RNAs is associated with erythrocyte Ro ribonucleoproteins. EMBO J 1990;9(11):3683-9. [PMID: 1698620 DOI: 10.1002/j.1460-2075.1990.tb07580.x]
- Wolin SL, Steitz JA. Genes for two small cytoplasmic Ro RNAs are adjacent and appear to be single-copy in the human genome. Cell 1983;32(3):735-44. [PMID: 6187471 DOI: 10.1016/0092-8674(83)90059-4]
- Farris AD, Puvion-Dutilleul F, Puvion E, Harley JB, Lee LA. The ultrastructural localization of 60-kDa Ro protein and human cytoplasmic RNAs: association with novel electron-dense bodies. Proc Natl Acad Sci USA 1997;94(7):3040-5. [PMID: 9096342 DOI: 10.1073/pnas.94.7.3040]
- Gendron M, Roberge D, Boire G. Heterogeneity of human Ro ribonucleoproteins (RNPS): nuclear retention of Ro RNPS containing the human hY5 RNA in human and mouse cells. Clin Exp Immunol 2001;125(1):162-8. [PMID: 11472440 DOI: 10.1046/j.1365-2249.2001.01566.x]
- Chen X, Quinn AM, Wolin SL. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev 2000;14(7):777-82. [PMID: 10766734]
- Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, et al . The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell 2009;20(5):1555-64. [PMID: 19116308 DOI: 10.1091/mbc.e08-11-1094]
- Klasic M, Kristic J, Korac P, Horvat T, Markulin D, et al. DNA hypomethylation upregulates expression of the MGAT3 gene in HepG2 cells and leads to changes in N-glycosylation of secreted glycoproteins. Sci Rep 2016;6:24363. [PMID: 27073020 DOI: 10.1038/srep24363]
- Anugraham M, Jacob F, Nixdorf S, Everest-Dass AV, Heinzelmann-Schwarz V, et al . Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: glycan structures reflect gene expression and DNA methylation status. Mol Cell Proteomics 2014;13(9):2213-32. [PMID: 24855066 DOI: 10.1074/mcp.M113.037085]
- Horvat T, Muzinic A, Barisic D, Bosnar MH, Zoldos V. Epigenetic modulation of the HeLa cell membrane N-glycome. Biochim Biophys Acta 2012;1820(9):1412-9. [PMID: 22192783 DOI: 10.1016/j.bbagen.2011.12.007]
- Wahl A, Kasela S, Carnero-Montoro E, van Iterson M, Stambuk J, et al. IgG glycosylation and DNA methylation are interconnected with smoking. Biochim Biophys Acta Gen Subj 2018;1862(3):637-48. [PMID: 29055820 DOI: 10.1016/j.bbagen.2017.10.012]
- Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc Natl Acad Sci USA. 2003;100:14846-51. [PMID: 14657396 DOI: 10.1073/pnas.2335201100]
- Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science 2000;287(5460):2007-10. [PMID: 10720325 DOI: 10.1126/science.287.5460.2007]
- Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, Bertozzi CR. Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat Methods 2015;12(6):561-7. [PMID: 25894945 DOI: 10.1038/nmeth.3366]
- Gaziel-Sovran A, Hernando E. miRNA-mediated GALNT modulation of invasion and immune suppression: a sweet deal for metastatic cells. Oncoimmunology 2012;1(5):746-8. [PMID: 22934269 DOI: 10.4161/onci.19535]
- Li Y, Zeng C, Hu J, Pan Y, Shan Y, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol 2018;11(1):89. [PMID: 29970122 DOI: 10.1186/s13045-018-0632-2]
- Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer 2004;4(1):45-60. [PMID: 14681689 DOI: 10.1038/nrc1251]
- Xu J, Xiao Y, Liu B, Pan S, Liu Q, et al. Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway. J Exp Clin Cancer Res 2020;39(1):54. [PMID: 32209115 DOI: 10.1186/s13046-020-01562-6]
- Pan S, Liu Y, Liu Q, Xiao Y, Liu B, et al. HOTAIR/miR-326/FUT6 axis facilitates colorectal cancer progression through regulating fucosylation of CD44 via PI3K/AKT/mTOR pathway. Biochim Biophys Acta Mol Cell Res 2019;1866(5):750-60. [PMID: 30742932 DOI: 10.1016/j.bbamcr.2019.02.004]
- Li Y, Sun Z, Liu B, Shan Y, Zhao L, et al . Tumor-suppressive miR-26a and miR-26b inhibit cell aggressiveness by regulating FUT4 in colorectal cancer. Cell Death Dis 2017;8(6):e2892. [PMID: 28640257 DOI: 10.1038/cddis.2017.281]
- Liang L, Gao C, Li Y, Sun M, Xu J, et al. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis 2017;8(8):e2968. [PMID: 28771224 DOI: 10.1038/cddis.2017.352]
- Liu Q, Ma H, Sun X, Liu B, Xiao Y, et al. The regulatory ZFAS1/miR-150/ST6GAL1 crosstalk modulates sialylation of EGFR via PI3K/Akt pathway in T-cell acute lymphoblastic leukemia. J Exp Clin Cancer Res 2019;38(1):199. [PMID: 31419997 DOI: 10.1186/s13046-019-1365-y]
- Liu B, Liu Q, Pan S, Huang Y, Qi Y, et al. The HOTAIR/miR-214/ST6GAL1 crosstalk modulates colorectal cancer procession through mediating sialylated c-Met via JAK2/STAT3 cascade. J Exp Clin Cancer Res 2019;38(1):455. [PMID: 31694696 DOI: 10.1186/s13046-019-1468-5]